HPR volume 9 issue 44: news and travel (11 December)
Updated 29 December 2015
1. Surgical site infection surveillance in NHS hospitals annual report in summary
PHE will shortly publish the latest results from its surveillance of surgical site infections (SSI) programme, summarising data collected by 232 NHS hospitals and independent NHS Treatment centres in England between April 2010 and March 2015 [1]. The report presents the rate of SSIs for 17 categories of surgical procedures based on infections detected during hospital patients’ post-operative stays (“inpatient SSIs”) combined with infections detected on re-admission after initial discharge (“re-admission SSIs”). Re-admission surveillance became a requirement of the Surgical Site Infection Surveillance Service (SSISS) programme from July 2008; prior to this re-admission cases were collected on a voluntary basis.
NHS Trusts in England performing orthopaedic surgery in one of the four mandatory surveillance categories (hip prosthesis, knee prosthesis, reduction of long bone fracture and repair of neck of femur) are required to undertake SSI surveillance in at least one of these surgical categories in one hospital for a minimum of one quarter per financial year. NHS Trusts also have the option of participating in any of the additional 13 surgical categories included in the national surveillance scheme covering gastro-intestinal, cardiothoracic, neurosurgery, obstetrics and gynaecology, vascular and other general surgery procedures.
Surveillance of SSIs is undertaken by hospitals in England using standard definitions for infections that affect the superficial incisional site, the deeper layers or those involving the joint or organ-space as outlined in the surveillance protocol [2]. Patients are systematically, prospectively followed-up to identify infections occurring within 30 days of surgery or within one year if a prosthetic implant is used.
The report describes hospital participation in surveillance over time, data quality indicators, trends and risk factors for SSI. An accompanying supplement lists orthopaedic SSI rates by named NHS Trust [3], also available in due course from the NHS Choices website.
1.1 Trust participation
In 2014/15, 138 NHS Trusts and an additional eight NHS Treatment centres participated in the mandatory orthopaedic surveillance, contributing data on 102,496 procedures. Four eligible NHS Trusts did not participate. Of those that participated, eight NHS Trusts were identified as high outliers with an incidence of SSI higher than expected. An additional eight (including one NHS treatment centre) were identified as low outliers. All 16 providers have been contacted and encouraged to review their clinical and surveillance practices.
1.2 Key findings
Key findings are:
-
between 2008/09 and 2014/15, a significant decrease in the inpatient / re-admission detected SSI incidence occurred for repair of neck of femur, reaching 1% in 2014/15. No overall trends for hip or knee prosthesis were found with the incidence remaining low (<1%) for both categories or for reduction of long bone fracture with an incidence of (1.4%)
-
a large and significantly decreasing trend was found for gastric surgery with an SSI rate of 1.6% in 2014/15.In contrast, a small but significantly increasing trend in SSI was found for patients undergoing spinal surgery with an SSI rate of 1.3% in 2014/15
-
Staphylococcus Aureus accounted for 13% of inpatient SSIs in 2014/15 following a decreasing trend since 2006/7 due to decreases in methicillin-resistant S. aureus (MRSA) which now account for 3% of SSI. Enterobacteriaceae increased from 2008/9 and accounted for 25% of SSIs in 2014/15
-
S. Aureus was the predominant organism in orthopaedic and spinal surgery accounting for ≥36% of cases in 2014/15. Coagulase-negative staphylococci were the predominant causes for coronary artery bypass graft infections and enterobacteriaceae for large bowel surgery.
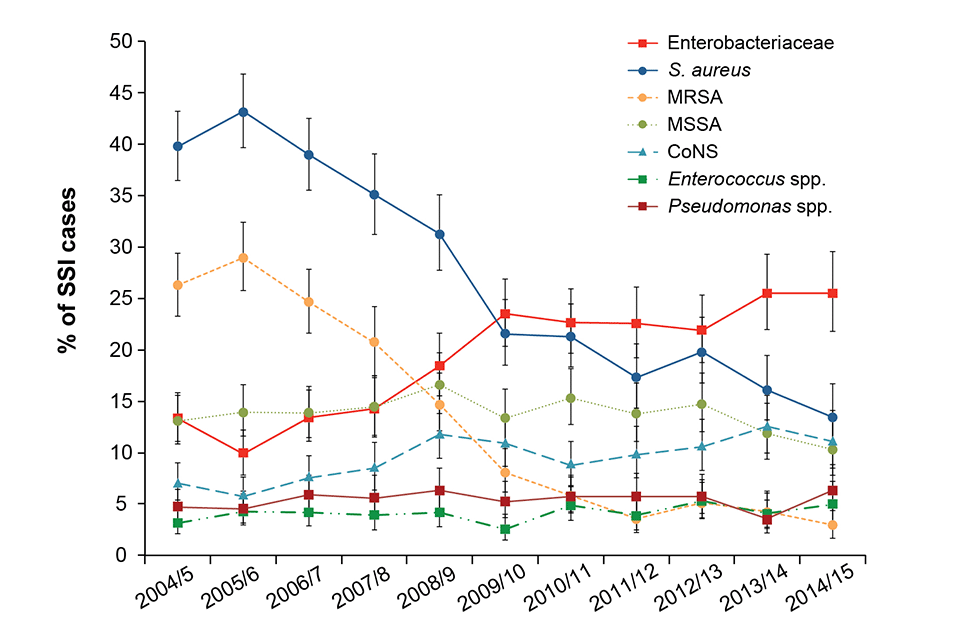
Trends in key micro-organisms reported as causing inpatient SSIs in NHS hospitals, England (excluding breast, cardiac (non-CABG), cranial, and spinal surgery)
1.3 References
-
‘Surveillance of Surgical Site Infections in NHS Hospitals in England 2014/15’. See: Surgical site infections (SSI) surveillance: NHS hospitals in England.
-
PHE (2013). Protocol for the surveillance of surgical site infection. Version 6 June 2013.
-
‘Surgical Site Infections surveillance: NHS Trust tables 2014/15’. See: Surgical site infections (SSI) surveillance: NHS hospitals in England.
2. Zika virus in the Americas and increase in microcephaly
Following the emergence of Zika virus (ZIKV) in the Pacific in 2014 [1], large outbreaks have been reported in Brazil and Colombia during 2015 resulting in spread to several other countries in South and Central America.
ZIKV infections were first reported in Easter Island (a Chilean island in the south eastern Pacific Ocean) in February 2014 [1]. The first autochthonous cases in mainland South America were reported in Brazil in May 2015 with 18 states affected as of 1 December 2015 [2]. To date, locally acquired cases have been reported in Brazil, Colombia, El Salvador, Guatemala, Mexico, Panama, Paraguay, Suriname and Venezuela. In addition, in 2015, cases have also been reported in Cape Verde [1].
ZIKV usually causes a mild febrile illness similar to dengue and, like dengue, is transmitted by Aedes mosquitoes which are present throughout South and Central America and the Caribbean. It is likely that further cases will be reported in other countries in the Americas in coming weeks/months.
In November 2015, the Brazilian Ministry of Health (MoH) reported a 20-fold increase in the number of babies born with microcephaly [2]. As of 5 December 1,761 cases of microcephaly including 19 deaths had been reported across 14 states in Brazil [3] compared to the expected 150-200 cases per year that were reported 2010 to 2014. A possible relationship between the increase in microcephaly and the ongoing ZIKV outbreak has been suggested by the MoH although further investigations are ongoing to establish whether there is a causal relationship.
In addition to this, an increase of central nervous system malformations in foetuses and new-borns has been reported in French Polynesia following an epidemic of ZIKV infection. At least 17 such cases were reported during 2014–2015, coinciding with the ZIKV outbreaks on the French Polynesian islands. Of four women who were tested, all had detectable IgG antibodies to flavivirus; further tests are ongoing.
During the same ZIKV outbreak in French Polynesia, 37 cases of Guillain-Barré syndrome were also reported in patients who had presented with a previous viral syndrome. In Brazil, 121 cases of neurological manifestations and Guillain-Barré were notified between January and July 2015 in individuals who had a history of rash illness. Investigations are still ongoing in Brazil and French Polynesia to establish if Zika virus infection increases the risk of developing Guillain–Barré syndrome [1].
As a precaution, the National Travel Health Network and Centre (NaTHNaC) has reviewed and updated its advice for travellers to the Americas, particular for pregnant women [4, 5]. There is currently no vaccine available to prevent ZIKV and prevention of ZIKV relies on avoiding mosquito bites [6].
Health professionals should consider ZIKV among the differential diagnoses of patients with fever returning from the Americas. If a case of ZIKV is suspected, appropriate samples for testing (together with a full travel and clinical history with relevant dates) should be sent as early as possible to the Public Health England, Rare and Imported Pathogens Laboratory.
The Imported Fever Service is available to local infectious disease physicians or microbiologists should specialist advice be needed on 0844 7788990.
Health professionals should also be vigilant for any increase of neurological and autoimmune syndromes (in adults and children), or congenital malformations in new born infants (where the cause is not otherwise evident) in patients with a history of travel to areas where ZIKV transmission is known to occur [4].
Further information on the PHE Zika virus health protection guidance webpage.
2.1 References
-
ECDC (10 December 2015). Rapid Risk Assessment: Zika virus epidemic in the Americas: potential association with microcephaly and Guillain-Barré syndrome.
-
Pan American Health Organization (1 December 2015). Epidemiological Alert: Neurological syndrome, congenital malformations, and Zika virus infection. Implications for public health in the Americas.
-
Ministry of Health, Brazil (8 December 2015). Boletim Epidemiológico: Ministério da Saúde divulga novos casos de microcefalia (in Portuguese).
-
National Travel Health Network and Centre (7 December 2015). Zika virus in the Americas: update and advice for pregnant women.
-
National Travel Health Network and Centre. Pregnancy factsheet.
-
National Travel Health Network and Centre. Insect and tick bite avoidance factsheet.
3. Study on use of WGS in TB diagnostics and drug susceptibility testing
The feasibility of using whole genome sequencing as a routine diagnostic method to underpin public health measures to control tuberculosis has been demonstrated in an international study that compared the performance of the technique – in terms of cost, speed and discriminatory power – with traditional “wet” laboratory methods [1].
The study – a pilot involving eight laboratories in Europe and North America – compared, in respect of testing of all newly positive mycobacterium cultures over a seven month period in 2013/14, the results obtained by sequencing as against those obtained by traditional culture-based methods. In each case, mycobacteria present were identified and, for tuberculosis, resistance to first-line and second-line antibiotics predicted.
The main conclusions were that cost should not be an obstacle to the implementation of WGS for TB surveillance; also that MTBC identification and drug susceptibility test results were available significantly faster than when traditional methods were used. WGS also offered the ability to monitor emergence of new resistance mechanisms, and higher-resolution outbreak monitoring, on a timescale weeks faster than with traditional diagnostics [2].
Public Health England is now assessing whether it is practicable for WGS to replace traditional laboratory test methods for all routine MTBC testing. That would be the second example in recent months of WGS becoming the default technology for surveillance of a significant category of communicable disease, following the recent replacement of traditional phage typing and “first generation” molecular typing (MLVA and PFGE) techniques by WGS in salmonella outbreak surveillance [3].
Molecular biology techniques have to date been applied to complement, rather than replace, traditional culture-based laboratory methods. For the control of HCAIs, for example, the discriminatory power of the MLVA technique has facilitated identification and analysis of Clostridium Difficile infection cases and clusters, helping to confirm or refute suspected case clusters [4].
In the case of public health measures employed to prevent and control tuberculosis, a MIRU-VNTR service has been provided by PHE to Consultants in Communicable Disease Control, NHS microbiologists and consultant chest /infectious disease physicians throughout England since 2005, playing a significant role in accelerating traditional outbreak/cluster investigation procedures, either to reveal or confirm the existence of epidemiological links between cases or to demonstrate that clusters previously presumed to have been the result of transmission events were not in fact connected (particularly important in nosocomial settings).
In the past, however, the costs associated with WGS – both in terms of hardware, paucity of personnel qualified to implement the technology and bioinformatics systems/facilities – have been a significant obstacle to further expansion.
The study’s conclusion on the competitive cost of WGS acknowledges that this currently only applies in high-income countries where the necessary sequencing and bioinformatics resources are available (the pilot involved a “decentralised-sequencing, centralised-analysis” configuration of resources). However, it is noted that as WGS technology continues to develop and portable systems become available, WGS will also revolutionise diagnostics in low-income countries.
3.1 References
-
Pankhurst LJ, del Ojo Elias C, Votintseva AA, Walker TM, Cole K, Davies J, et al (2015).Rapid, comprehensive, and affordable mycobacterial diagnosis with whole genome sequencing: a prospective study Lancet Respiratory Medicine (published online 3 December).
-
‘Scientists use DNA technology to diagnose cases of TB faster’. PHE press release, 3 December 2015.
-
Use of genomics in salmonella surveillance. HPR 9(40), 13 November 2015.
-
Leeds CDRN study on use of genetic fingerprinting in CDI case-cluster and outbreak investigation. HPR 6(1), 6 January 2012.
