Genomics Beyond Health - full report (accessible webpage)
Published 26 January 2022
What could genomics mean for wider government?
-
What is the genome?
-
Current applications of Genomic data.
-
Genomic analysis and predication traits.
-
What could genomics tell us about people I the future?
-
Genomic data, risks, privacy and security
-
What could genomics mean for wider government?
Preface

DNA, the foundation of all biological life, was first discovered in 1869 by Swiss chemist Friedrich Miescher. A century of gradual discoveries allowed James Watson, Francis Crick, Rosalind Franklin, and Maurice Wilkins to deduce the now famous ‘double-helix’ model in 1953, two bonded chains that coil around each other. With the structure of DNA finally understood, it was a further 50 years until the full human genome was sequenced in 2003 by the Human Genome Project.
The sequencing of the human genome at the turn of the millennium was a critical point in our understanding of human biology. Finally, we could read nature’s genetic blueprint.
Since then, the technologies with which we can read the human genome have developed at a rapid pace. The first genome took 13 years to sequence, meaning that many scientific studies were able to look at only specific sections of DNA. Now, sequencing a whole human genome can be completed in a single day. This technological advance in sequencing presents a step change in our ability to understand the human genome. Large scale scientific studies have improved our understanding of how specific sections of DNA – genes – relate to some of our traits and characteristics. However, the effects that genes have on different traits is a very complicated puzzle; each of us has around 20,000 genes which operate in complex networks to influence our traits.
The major focus of scientific research so far has been on health and disease, where we have made remarkable progress for some conditions. Here, genomics is becoming a fundamental tool in our understanding of health and the progression of disease. The world leading genomic infrastructure that the UK has developed has placed it at the head of genomic data capacity and research internationally.
This has been clear throughout the COVID pandemic, where the UK has led efforts in sequencing the genome of the SARS-CoV-2 virus. Genomics is expected to form a central pillar of future UK health service delivery by the NHS. It should increasingly offer earlier identification of disease, diagnosis of rare genetic disease, and help to better tailor people’s healthcare.
Scientists are developing a better understanding of how our DNA relates to a broad range of traits in fields beyond health, such as employment, sport, and education. This research has been able to capitalise on the genomic infrastructure that was developed for health research; transforming our understanding of the way that a broad range of human characteristics are formed and developed. While our genomic knowledge of non-health traits is growing, it lags well behind that of health traits.
The opportunities and challenges we see playing out in health genomics - such as the need for genetic counselling or questions around when a test is informative enough to justify use - provide a window into the potential future of non-health genomics.
In addition to growing use in the health sector, more of the public are gaining exposure to genomic knowledge through private companies that offer direct-to-consumer services. For a fee, these companies offer people the opportunity to explore their ancestry and gain genomic insights about a range of characteristics.
The increasing knowledge being developed through international research has allowed successful development of new technologies, and the accuracy with which we can predict people’s characteristics from their DNA is improving. Beyond understanding, it is now technologically possible to edit specific genes.
While genomics has the capacity to be transformative for many aspects of society, there are ethical, data and security risks that may accompany its use. Nationally and internationally, the use of genomics is governed by a mix of voluntary codes and more general regulations not specially designed for genomics, such as general data protection laws. As the power of genomics increases, and uses proliferate, government will increasingly face choices about whether this approach still ensures the safe integration of genomics into society. Capitalising on the UK’s various strengths in genomic infrastructure and research will require a coordinated effort across government and industry.
Sir Patrick Vallance
Government Chief Scientific Adviser
Introduction and overview
How would you feel if your genomic information made your car insurance more expensive?
If you could find out whether your child was likely to excel at sport or academic pursuits, would you?
Should criminal sentencing account for a person’s genomic predispositions?
These are just some of the questions that we might face in the not-too-distant future as genomic science provides us with more and more information about the human genome, and the role it plays in influencing our traits and behaviours.
A person’s genomic information - their unique deoxyribonucleic acid (DNA) sequence - can already be used to make some medical diagnoses and personalise their treatment. But we are also beginning to understand how the genome can influence people’s traits and behaviours beyond health.
There is already evidence that non-health traits such as risk taking, educational attainment, and substance abuse are influenced by the genome. As we understand more about how genes influence traits, we may be better able to predict how likely and to what extent someone will develop these traits from their genomic sequence.
This raises several important questions. How might this information be used? What could this mean for our society? And how might policy across departments need to adapt? Will we need more regulation? How will we manage the ethical questions raised, address the risk of discrimination, and the potential threats to privacy?
While some of the potential uses of genomics may not be realised in the short or even medium-term, people are already exploring new ways to use genomic information today. This means that now is the time to anticipate how genomics might be used in the future. We should also consider the impact that genomic services might have if they are offered to the public before the science is truly ready. This will allow us to properly consider the opportunities and risks that these new applications of genomics may represent, and to identify actions we can take in response.
This report introduces genomics for the non-specialist, explores how the science is developing, and tries to consider the implications across a variety of sectors. The report looks at what is possible now, what might be possible in the future, and explores where the capabilities of genomics are potentially being oversold.
The scope of this report
Genomics is not just a health policy issue. It could impact a huge variety of policy areas, from education and criminal justice to employment and insurance. This report focuses on human genomics outside the health sphere. It also explores genomic applications in agriculture, ecology, and synthetic biology, to provide a sense of the breadth of its potential use in other sectors.
However, much of our knowledge about human genomics comes from studies examining its role in health and disease. Health is also where many potential applications have been developed. So that’s where we start too, with Chapters 2 and 3 providing a background to the science and development of genomics. This provides context for the field of genomics and the technical knowledge necessary to understand how genomics could impact sectors beyond health. Readers who don’t need the technical background can safely jump from this introduction to Chapters 4, 5 and 6, which provide the substance of this report.
What is genomics?
Humans have long been fascinated by our genetics, and the role they play in making us who we are. We are eager to understand how hereditary factors influence our physical features, health, personalities, characteristics, and skills, as well as how they interact with environmental influences.
£4 billion, 13 years Cost and time it took to draft sequence the first human genome (cost adjusted for inflation).
Genomics is the study of an organism’s genome – their entire DNA sequence, and how all our genes work together in our biological system. In the 20th century, studies on the genome were often limited to the observation of twins to explore the role of genetics vs. environment in physical and behavioural characteristics (or ‘nature vs. nurture’). However, the mid 2000s marked the first publication of the human genome and the development of faster and cheaper genomic technologies.
These technologies meant that researchers could finally investigate the genetic code directly, and at a fraction of the time and cost as was previously possible. Sequencing the whole human genome, which once took years and cost billions of pounds, now takes less than a day and costs about £800[footnote 1]. Researchers can now analyse the genomes of hundreds of individuals or draw on biobanks containing the genomic information of many thousands more. As a result, genomic data is being accumulated in vast quantities for research use.
Until now, genomics has mostly been utilised within healthcare and medical research. For example, identifying the presence of a faulty gene variant, such as the BRCA1 variant that is associated with breast cancer. This can allow earlier preventative treatment that would not be possible without genomic knowledge. However, as our understanding of genomics grows, it’s becoming clear that the influences of the genome extend well beyond health and disease.
What is DNA?
The quest to understand our genetic makeup has taken huge steps forward over the past 20 years. We are beginning to understand the genome’s structure and function, but there is a lot left to learn.
3.2 billion Approximate number of letters (bases) in the human genome.
We have known since the 1950’s that our DNA sequence is a code, and this code contains the instruction for our cells on how to make proteins. Each gene corresponds to a different protein, and the proteins that are made give rise to an organism’s traits (such as eye colour, or flower size). DNA can influence traits through a variety of mechanisms; a single gene might determine a trait (like ABO blood group), several genes may work together synergistically (as seen with height and skin pigmentation), or some genes may override one another, masking the influence of others (like baldness and hair colour).
Most traits are influenced by many (likely thousands) of different segments of DNA working together. But mutations to our DNA can cause the proteins to change – this might then cause the trait to change. This is the primary driving force behind biological variability, diversity, and disease. Mutations can confer advantages or disadvantages to an individual, a neutral change, or they may have no effect at all. They can be passed down through families or occur from the point of conception. However, if they occur during adulthood, this generally limits their effects to the individual rather than their offspring.
The variability of traits can also be influenced by epigenetic mechanisms. These can control whether genes are switched on or off. Unlike genetic mutations, they are reversible and partly influenced by our environment. This means that understanding the cause of a trait is not a simple study of what genetic sequence influences each trait. It is necessary to consider genetics within a wider context, with an appreciation of the networks and interactions across the whole genome, and the role of the environment.
Genomic technologies
Genomic technologies can be used to determine someone’s genetic sequence. These techniques are now used widely in many research studies and are increasingly offered by commercial companies for the purposes of health or ancestry analysis. The methods by which a company or study will determine someone’s genetic sequence varies, but until recently most have used a technique called DNA microarray. Microarrays measure sections of a person’s genome rather than reading the entire sequence. Historically, microarrays have been easier, quicker, and cheaper to use than other methods, but their use comes with some limitations.
Once data has been accumulated, it can be studied at scale through a genome wide association study (or GWAS). These studies look for gene variants which are associated with a specific trait. However, to date, even the largest studies have only been able to identify a fraction of the genetic effects behind many traits compared to what we expect from twin studies. The inability to identify all the relevant genetic markers for a trait is known as the “missing heritability” problem.[footnote 2]
However, the power of GWAS to identify relevant gene variants improves with the addition of more data, so the missing heritability problem may be solved as increasing amounts of genomic data are collected.
Furthermore, as costs continue to fall and the technology continues to improve, more researchers are using a technique called whole genome sequencing instead of microarrays. This reads the whole genomic sequence directly rather than parts of it. Sequencing can overcome many of the limitations associated with microarrays, so the data obtained is richer and more informative. This data also helps to reduce the missing heritability problem, and this means that we are beginning to understand more about which genes work together to influence a trait.
97.7% Decrease in cost to sequence a human genome since 2010.
Similarly, the massive collection of whole genome sequences now planned for healthcare purposes will provide a much richer and more powerful research dataset. This will benefit those studying both health and non-health traits.
As we develop an understanding of how genes influence traits, we can better predict how different genes may act together on a specific trait. This is done by combining estimated effects from multiple genes into a single measure of genetic liability, known as polygenic scoring. Polygenic scores tend to provide more accurate predictions for how likely an individual is to develop a trait than are possible from individual genetic markers.
Polygenic scores are now becoming popular in health research, with the objective of one day using them to guide clinical interventions at an individual level. However, polygenic scores are constrained by GWAS and as such many do not yet predict their target trait very accurately, with a polygenic score for height only achieving a predictive accuracy of 25 percent.[footnote 3] This means that for some traits they may be less accurate to other diagnostic methods, such as a blood test or MRI scan. Nevertheless, as genomic data improves, so should the accuracy of polygenic scores. In the future, polygenic scoring could provide clinical risk information far earlier than is possible with traditional diagnostic tools, and they may also be used to predict non-health traits in the same way.
But as with any methodology, there are limitations. The key limitation of a GWAS is the diversity of the data it uses, which has not reflected the diversity of the general population. Research has shown that as much as 83 percent of GWAS have been conducted on cohorts entirely of European ancestry.[footnote 4] This is clearly problematic because it means that a GWAS may only be relevant for a select population. Developing and using predictive tests based on findings from population biased GWAS may therefore be discriminatory to those outside the GWAS population.
0.3% Proportion of GWAS which have been based on data from people of African ancestry.
For non-health traits, predictions from polygenic scores are currently less informative compared to non-genomic information that is already available. For example, a polygenic score for predicting educational attainment (currently one of the best performing polygenic scores available) is less informative than a simple measure of parental education.[footnote 5] The predictive power of polygenic scores will inevitably increase with larger, more diverse studies, and studies based on whole genome sequencing data.
The current uses of genomics
Health
Genomic research has focused heavily on the genomics of health and disease, helping to identify sections of the genome that influence disease risk. Our knowledge about the role of genomics differs across diseases. For some diseases that are caused by a single gene, such as Huntington’s, we can accurately predict an individual’s likelihood of developing the disease from their genomic data. For diseases that are caused by many genes in combination with environmental exposures, such as coronary heart disease, genomic prediction is far less accurate. Generally, the more complex a disease or trait is, the more difficult it is to understand and predict accurately. However, predictive accuracy is improving as study cohorts become larger and more diverse.
The UK is at the forefront of health genomics research. We have already developed large scale infrastructure in genomic technologies, research databases, and computational power. The UK has contributed widely to global genomic knowledge, particularly during the COVID-19 pandemic, where we have led efforts to sequence the genome of the SARS-CoV-2 virus and its emerging variants.
Genome UK is the UK’s ambitious genomic healthcare strategy, which will see the NHS integrate genomic sequencing into routine clinical care to diagnose rare diseases, cancer, or infectious disease.[footnote 6]
It will also lead to a massive increase in the number of human genomes available for research. This should allow for wider studies and will unlock further applications for genomics. As a world leader in the development of genomic data and infrastructure, the UK could lead internationally on the ethics and regulation of genomic science.
1,000,000 by 2024
Whole genome sequencing target of GMS and UK BioBank.
Direct-to-consumer testing:
Direct-to-consumer (DTC) genetic testing kits are marketed and sold directly to consumers without the involvement of a healthcare provider. A swab of saliva is sent off for analysis, providing the consumer with personalised health or ancestry analysis in just a few weeks. This market is growing rapidly, and tens of millions of consumers across the world have submitted DNA samples for commercial sequencing, to gain insights into their health, ancestry, and genetic predisposition to traits.
While rapidly growing in popularity , there are some risks associated with DTC testing.[footnote 7]
The accuracy of some genomic-based insights which underpin direct- to-consumer services can be very low. The tests may also impact an individual’s privacy, through data sharing, identifiability of relatives, and potential lapses in cybersecurity protocols. Customers may not fully understand these issues when they engage with a DTC testing company.
$2.7 billion by 2025
Predicted growth of the global DTC testing market (USD).
DTC genomic tests for non-medical traits are also largely unregulated. They fall beyond the scope of legislation covering medical genomic tests, relying instead on voluntary self-regulation by the test providers. Many of these companies are also based outside of the UK and are not subject to UK regulation.
Forensics:
DNA sequences are uniquely powerful in forensic science for the identification of unknown individuals. Basic DNA analysis has been widely used since the invention of DNA fingerprinting in 1984, and the UK’s National DNA Database (NDNAD) holds profiles of 5.7 million individuals and 631,000 crime scene records.[footnote 8]
The database may only be used for the detection or prevention of crime, or to identify bodies following deaths from natural disasters.
DNA fingerprinting counts the number of repeated sequences in pre-determined short tandem repeat (STR) areas of the genome to differentiate between individuals, rather than matching the sequences directly. This allows suspects to be matched to samples taken from crime scenes. The forensic DNA profile used by police in England and Wales, DNA-17, uses 16 different STR areas (or loci) for comparison. This means that the probability of two full DNA profiles from unrelated individuals matching by chance is around one in a billion.
In the future, targeted or full genome sequencing could replace the STR techniques currently in use. This could enable DNA phenotyping (like a ‘predictive photofit’ image to predict the physical features of an unknown individual) or age prediction, alongside traditional DNA fingerprinting. But expanding the use of DNA in forensics, especially in a genomic capacity, raises issues around surveillance, privacy, and the potential for discrimination against certain groups.
0.78%
Average amount of DNA shared between third cousins.
Large commercial genomic databases may also be used in a forensic capacity. Some DTC companies provide consumers with a copy of their raw genomic data. The consumer may then choose to upload the data to a third-party genomic database for additional health, ancestry, or wellness analysis. US law enforcement agencies have used these databases to identify human remains or criminal suspects through matching samples to distant relatives, which is possible as far as a third cousin match.[footnote 9][footnote 10]
Non-human genomics:
Genomic science isn’t an exclusively human science – a point clearly demonstrated by the pivotal role that genomics has played in the response to COVID-19. In many ways, the non-human applications of genomics are even more advanced than the human applications. For example, genomics is often used to inform selective breeding processes in the agricultural sector, the development of genetically edited or modified crops or livestock, environmental monitoring for biological pathogens or indicators of pollutants, species cataloguing, or the modelling of species adaptation in response to climate change. Understanding more about how the genome relates to biological traits may also be used to inform the development of edited or completely synthetic genomes or biological life forms. These may themselves have a diverse spectrum of uses ranging from biomaterials and biofuels to new types of medicine and biological computers or sensors.
Whilst our report is focussed on human genomics, we also discuss a number of these applications in more detail to illustrate the breadth of scope. However, this is by no means exhaustive, and the range of applications is likely to increase as the science develops.
>90%
Percentage of soy, cotton and corn grown in the US (2018) which were genetically modified.
66,000
Number of species in the British Isles which are being sequenced by the Darwin Tree of Life Project.
2045
Prediction for when we’ll be able to build a full replica human genome synthetically (based on current progress).
The future uses of genomics: beyond health
Studies examining the role of genomics in health and disease have generated a wealth of knowledge about the fundamental workings of our genome. The range of fields beyond health, where this knowledge could be applied, is also growing. For example, genomic research into education has provided knowledge of the role that genomics plays in educational outcomes and how people learn.
Like many health traits, non-health traits are usually very complex, being influenced by thousands of genes and an array of environmental factors. This complexity, combined with the infancy of genomic research beyond health, means that understanding of how genomics applies to non-health fields lags behind its use in health. In many non-health fields, the operational implications of genomic prediction are also more challenging. For example, as non-health traits are especially complex, accurately predicting them is more difficult compared to traits which are influenced by one or a few genes.
Non-health genomic applications can therefore be grouped into two areas:
-
Those that are theoretically possible.
-
Those that are technically possible, given our current knowledge
Policymakers and practitioners will need to react to emerging developments in genomic science in two ways. First, to make the best use of developments in genomics they will need to address the technical and ethical issues that they raise. Second, policymakers will need to consider what mitigations and support should be in place to help customers and service providers navigate the genomic marketplace. The DTC genomic testing market is a useful example; it is an international market, mostly governed by voluntary codes, and tests may be offered to customers before they are fully supported by the evidence base, without this being made clear to the customer.
The limitations of the current evidence base should also be acknowledged. The underrepresentation of genomes from people of non-European ancestry in genomic databases impacts the accuracy of predictions that are made to all citizens made using that data. This must be addressed before genomics is used to predict behaviours and prescribe interventions, or we risk entrenching inequalities in key areas of life, such as in employment or education.
Regulation on the use of genomic technologies in non-health fields is patchy, and risks being outpaced by advances in the technology. Proactive regulation might prevent genomic technologies from being misused in non-health fields.
Employment
Genomic tests are currently used in very limited circumstances in employment, such as occasionally in professional sport. However, the rapid development of genomic science could prompt employers to use genomics more widely. This could potentially be in the selection of workers of optimal health or personality for a role, or to prevent workplace injury.
21-51%
Heritability estimate of musical ability.
The potential use of genomics in assessing candidates’ personal suitability would be most controversial. Many personality traits are reasonably influenced by the genome (i.e., are heritable), including extraversion (53 percent heritable), neuroticism (41 percent), agreeableness (41 percent), conscientiousness (44 percent), and openness to experience (61 percent).[footnote 11]
Yet tests designed to predict these traits frequently encounter technical and ethical barriers, and easier methods of testing currently exist.
However, this may change as predictive capability improves, and the UK has no explicit legislation barring the use of genomic analysis in employment scenarios. On the other hand, some countries have implemented proactive legislation - the Genetic Information Non-discrimination Act (GINA), enacted in the US in 2008, prohibits the use of genomic information in job hiring, redundancy, placement, or promotion decisions.[footnote 12]
Sport
Genetic tests can currently be used to screen for health conditions that would pose a risk to athletes, and for sex verification purposes in international competition. Variants of certain genes (including ACTN3, ACE, GALNTL6 and EPOR) have been associated with elite athletic performance. One variant of the EPOR gene is associated with an elevated red blood cell count, generating 25 to 50% more red blood cells than usual, and contributing to improved athletic endurance.[footnote 13]
1.23x
Number of times more likely that endurance athletes have a GALNTL6 gene.
DTC genomic tests intended to identify athletic potential or inform training regimes currently exist. However, they are not regarded as accurate or useful by prominent sporting bodies.[footnote 14]
In the future, gene editing techniques could be used to potentially enhance the performance of people whose genome does not include advantageous gene variants. However, the World Anti-Doping Agency (WADA) has pre-emptively outlawed gene doping and is developing techniques to detect it.
Education
Over a thousand genes have been identified that relate to educational and cognitive outcomes. However, it is very difficult to accurately predict a given pupil’s educational performance using currently available polygenic scores.
Despite this, DTC genomic testing companies are expanding into education-relevant fields and marketing these tests to parents. Three DTC genomic testing providers were offering genetics-informed IQ tests from a saliva sample in 2018.[footnote 15]
It is not clear how much traction these tests will gain with parents, or what support teachers will need in response to parents using them.
1,100
Number of genes currently thought to influence educational attainment.
Used effectively, one benefit of using polygenic prediction is that genomic data can be measured at birth, before other data used by educators is available. This means that it could enable earlier interventions to improve educational outcomes. This could include identifying students in need of academic support, designing learning approaches, or helping pupils with learning disabilities. However, there are no regulations in the UK governing the use of genomics in education, and their use could lead to stigmatisation of pupils.
Criminal Justice:
Some gene variants have been associated with behaviours linked to criminal behaviour. Variants of the MAOA and CDH13 genes have been associated with aggressive behaviour, and whilst substance abuse is not always a crime, addictive behaviours can also be heritable: cannabis addiction, alcohol dependence and cocaine use disorders have heritability estimates of 51 to 59%, 48 to 66%, and 42 to 79% respectively.[footnote 16]
Developing polygenic scores for susceptibility to substance abuse is a real possibility.
Genomic data has been raised as a mitigating factor in a small number of criminal cases in other countries, and this may become more common as the genomic evidence base improves, but there is no precedent for it in the UK. Youth services, social services, or the police may seek to explore the use of genomic prediction to deter or divert those who may be predisposed to criminal behaviour. However, polygenic scores are only an estimate of the likelihood of a particular trait manifesting, and our genes are only one of many influences on behaviour, including criminal behaviours. It also conflicts with the presumption of innocence, which is fundamental to our system of justice.
5-10%
Number of criminals convicted for severely violent crimes in Finland who possess MAOA and CDH13 gene variants.
Insurance:
Insurers might seek to use polygenic scores for heritable behavioural characteristics (such as risk-taking behaviour) and physiological factors (such as susceptibility to injury), to inform insurance policies in the future. This could impact on individuals’ car, home or even holiday insurance policies. Improvements in the accuracy and specificity of polygenic scores could make them more useful for insurance, however these uses would have to overcome public resistance. The UK insurance industry currently follows a voluntary code setting strict limitations on the use of health-related genomic information in determining eligibility for insurance.
The increased availability of DTC genomic tests could also increase information asymmetry between insurers and their customers. If consumers are aware of their genomic predispositions, but do not have to declare these to their insurer, they may affect insurance companies’ ability to accurately assess risk and price their products appropriately. Research has predicted that critical illness claims could increase by an average of 26% if the use of genetic information in underwriting is not permitted, though this is the subject of some debate.
The increased availability of DTC genomic tests could also increase information asymmetry between insurers and their customers. If consumers are aware of their genomic predispositions, but do not have to declare these to their insurer, they may affect insurance companies’ ability to accurately assess risk and price their products appropriately. Research has predicted that critical illness claims could increase by an average of 26% if the use of genetic information in underwriting is not permitted , though this is the subject of some debate.[footnote 17]
The risks and opportunities of genomic data
Developments in genomics are changing how we conceptualise privacy and anonymity, with implications for data security in both research and commercial (i.e., DTC) genomic databases. Genomic data is valuable information so should be protected, and the risks to privacy are not limited to the individual; their immediate family and close relations may be affected by any disclosure, for example if they share the risk of a health condition which might affect their insurance. DTC genomic testing therefore poses privacy risks of which its customers might not be fully aware.
Research projects are seeking to maximise the utility of large genomic datasets whilst minimising the risk to individual privacy. They use a variety of approaches, including mediated access to data through dedicated portals, and data encryption schemes. The privacy and security measures employed by these databases represent best practice. Policymakers and other processors of genomic data could learn from this approach and improve the protection of UK citizens’ genomic data.
Despite the commendable approach of research databases to uphold privacy and security, they (along with third party genomic databases and DTC genomic companies) operate under a patchwork of regulations and laws, including those on consumer protection, data protection, the human tissue act, medical device regulations, and advertising guidelines.[footnote 18]
This complex situation does not provide clarity to the organisations that curate and run the databases on their obligations to protect this data, or to provide reassurance to people volunteering their genomic information.
Genetic material and genomic sequences represent useful intellectual property. However, the question of whether genes can or should be patented remains controversial. Companies contend that their genetic discoveries are valuable assets that should be protected, whilst others see this as a land grab for a natural resource. Possible impacts of patenting genetic material include limiting open research, and the potential for increased costs for medical tests. Policymakers should consider whether the current system encourages innovation amongst biopharmaceutical companies, or if enhanced patent rights might limit research as we understand more about the genome.
Public opinion is generally positive about the potential benefits of genomics, particularly around forensic science. However, there are some ‘red-lines’ where the public feel genomics could disadvantage vulnerable people, and are wary of private businesses accessing their genomic information.
Overview conclusion
Genomics is already part of our daily lives. The UK’s strength in genomics has been integral to our ability to monitor the transmission of COVID-19 and identify new variants of the virus. We are still in the infancy of understanding the complexity of genomic data, and its place within a rich context of social and environmental influences, but this very rapidly changing.
We have worked with over 30 subject and policy experts from within the science and technology sectors, academia, and across government to develop this report.
They have helped us to understand the current landscape of genomics, and the reality of what we can and cannot learn from genomic data, now and in the future. Their expertise has ensured that our report reflects the current evidence base on genomic science and its applications.
The rapid technological and scientific advances in genomic science mean that genomic data and its potential applications are increasingly relevant to policymakers. The public are also starting to gain exposure to genomics through the healthcare system, and through DTC genomics companies offering a range of genomic services. Given the increasing deployment of genomic science in many sectors, regulations need to keep pace with the science, particularly outside the healthcare sector.
Proactive policy on genomics across sectors may be needed to protect UK citizens’ privacy, anonymity, and the security of their genomic information. Applications of genomics are likely to proliferate as the science progresses and technological barriers are overcome. It is important that policy is informed by the ethical and legal challenges that may arise, and that the legal framework is able to respond to these developments.
As our understanding of genomic science improves, governments around the world will face three key decision points:
-
Whether and when it is appropriate to use genomics to inform policy or deliver services. Each decision will require careful consideration of the ethics and any unintended consequences that could arise. There is an overarching need to address the lack of non-European ancestry in genomic data, or risk entrenching or increasing structural inequalities.
-
Whether the complex mix of laws, regulations and voluntary codes that currently governs genomics outside of healthcare settings remains fit for purpose. As potential applications of genomics increase, with implications for more sectors and lives, this question will become urgent. A structured framework governing the collection and use of genomic data outside of health could help protect citizens and provide clarity and certainty to innovators. However, over-regulation risks stifling innovation, and striking a balance will be key to making the most of genomics.
-
How to bring the public into decisions about using genomics and supporting them to navigate the consumer market. If governments hope to make more use of genomic data, that will ultimately need the public’s consent. As the number of tests grow, so too will the potential consequences for citizens of nefarious uses of genomic information or misinterpreted results.
There are no certainties as to how genomic technologies will develop, or how people might attempt to use genomic information. However, the direction of travel is becoming clearer. Now is the time to consider what might be possible, and what actions government and the public could take to maximise the benefits and mitigate the risks of our growing knowledge in this field.
How to read this report
Why is so much of the report about health? Although the focus of this report is on genomics beyond health, at times during this report it is necessary to discuss the genomics of health. This is because our knowledge of genomics predominately comes from studies examining its role in health and disease, so many of the concepts, theories and terminology used in genomics have developed through this lens. However, if you want to understand the science of genomics, the mechanisms of genetic inheritance and the limitations of this kind of research, then read chapters 2 and 3. As the most advanced area, the health sector also gives us a model of how other sectors may adapt to and use advances in genomics, so we briefly discuss genomic medicine in chapter 4 in order to illustrate the revolutionary potential genomics may have on a sector when the science reaches maturity.
Isn’t this all speculation? It is important to recognise that some of the potential uses of genomics beyond health may not be realised in the short or even medium-term future. However, it is only by considering how people may apply, or attempt to apply the science, that policy and regulatory systems can stay ahead to maximise benefits and minimise risks. Given the speed at which the science and technology are developing, policymakers may need to start considering the impact on their sectors now.
There is a huge amount to cover and consider, not all of which can be included in this report. Rather, we hope that it provides the basis for discussion within departments and enables futures-focused exploration of potential scenarios.
Report structure
Whilst the report structure is sequential, it is designed such that readers can focus on specific sections that are relevant to them if they prefer. Each of the chapters is structured with its key points outlined first to aid understanding for those not familiar with the concepts discussed, and considerations for policymakers last.
The chapters are organised as follows:
-
Chapter 2 introduces the basics of DNA, describing the structure and function of the human genome, how mutations in our genetic code arise, and how genes influence traits.
-
Chapter 3 describes the jump from genetic to genomic science, explaining how technological advances have allowed us to sequence the entire human genome, the types of studies that are informing genomic knowledge, and the technical process by which genomics can be used to predict traits.
-
Chapter 4 explains how genomics is currently being used in society, in human-focussed applications and beyond. We discuss the use of genomics in medicine, direct-to-consumer testing, forensic science, synthetic biology, agriculture, food, and environment and ecology.
-
Chapter 5 explores how genomics may be used in the future across a range of human-specific fields beyond health. We cover the potential use of genomics in employment, sport, education, criminal justice, and insurance. We also discuss how advances in genome editing may interface with non-health traits in the future.
-
Chapter 6 outlines the risks, security and regulation surrounding genomic data both now and in the future. We discuss data protection and public attitudes to the use of genomics, highlighting issues of concern.
-
Chapter 7 concludes this report, highlighting the key areas for policymakers.
DNA: the basics
This chapter provides a general introduction to the fields of genetics and genomics with explanations of key concepts such as DNA, mutation, heritability, phenotypes, and epigenetics. A basic understanding of these concepts will aid understanding of how they determine both visible and invisible traits that can pass between generations and within populations.
Key messages:
-
Our DNA instructs our cells to make proteins, with each gene corresponding to a different type of protein. The proteins give rise to distinguishing features (i.e., traits), such as eye colour, height or flower pigmentation.
-
How DNA influences our traits occurs through a variety of mechanisms; a single gene might cause a trait (like a blood group), several genes may work together synergistically (like height and skin pigmentation), or some genes may override one another, masking the influence of others (like baldness and hair colour).
-
Most traits are influenced by many (likely thousands) of different segments of DNA working together synergistically.
-
But changes (mutations) to our DNA sequence can cause the proteins to change – this can then cause the trait to change, and this is the primary driving force behind biological variability, diversity, and disease.
-
Mutations can confer advantages or disadvantages to an individual, or they may have no effect at all. The term ‘mutation’ is therefore not always reflective of a disadvantageous change.
-
Mutations can be passed down through families or accumulate from the point of conception and throughout adulthood.
-
The variability of traits can also be influenced by epigenetic mechanisms. These can control whether genes are switched on or off, and unlike genetic mutations, they are reversible and partly influenced by our environment.
-
This means that understanding the cause of a trait is not a simple study of what genetic sequence causes each protein – rather, it is necessary to consider genetics within a wider context, with an appreciation of the networks and interactions across the whole genome.
What is DNA?
Our genetic material is stored inside our cells as chromosomes. Most humans have 23 pairs of chromosomes (46 in total), inheriting one copy of each chromosome from either parent. These 23 pairs consist of 22 autosomes, present in both male and female, and 1 pair of sex-specific chromosomes, known as X and Y. Generally, the inheritance of two X chromosomes produces a female, whereas inheritance of one X and one Y chromosome produces a male.
Our chromosomes are made of tightly packed bundles of deoxyribonucleic acid, DNA. At the most fundamental level, our DNA is shaped like a double helix, consisting of two coiled chains of nucleotides (also known as bases). These nucleotides are bonded together in the centre of the double helix in pairs, much like the rungs of a ladder.
The nucleotides which make up our DNA are simple molecules consisting of a carbon-based sugar backbone (black spirals, Figure 1) bonded to a base molecule consisting of nitrogen, oxygen and hydrogen. There are four types of nucleotide, defined by their differences in this base molecule. The four nucleotides are known as adenine (A), thymine (T), cytosine (C) and guanine (G), and are shown in Figure 1. Importantly, these bases are complementary, meaning that they only ever bond to their specified partners. In human DNA, the nucleotide pairs are adenine and thymine, and cytosine and guanine.
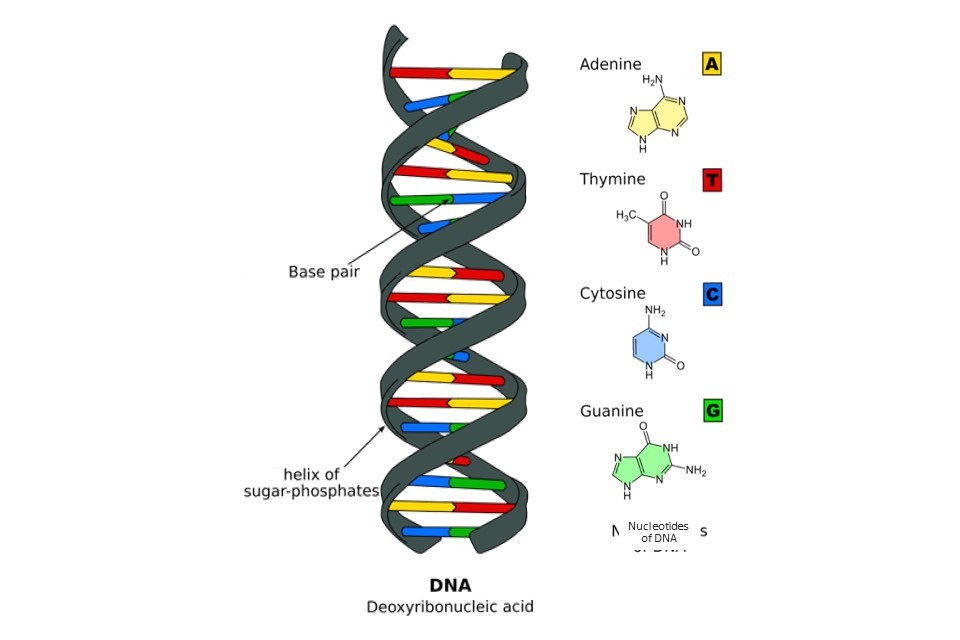
Figure 1: The essential structure of DNA, illustrating the nucleotides/bases spanning the centre of the double helix. Figure adapted under CreativeCommons from Wikimedia.
The function of DNA is to provide the instructions to make an end product, usually a protein. This is known as coding DNA, and sections of protein-coding DNA are known individually as genes. Alternatively, DNA can regulate these protein-coding DNA elements, where it is known as non-coding DNA. Together, these two categories of DNA constitute a person’s genome.
Current estimates suggest that the human genome, although 3.2 billion base pairs long, only contains approximately 20,000 protein-coding genes. This accounts for 1.5% of the total genome, leaving up to 98.5% of the genome having non-coding functions.[footnote 19] .
Presently, our understanding of protein-coding DNA far outweighs our knowledge of non-coding DNA, thus the focus of this report will be primarily on the role of protein-coding DNA, its influence on traits, and the predictive capabilities surrounding this.
The function of protein-coding DNA
Proteins are chains of amino acids. In protein-coding DNA, the order of nucleotides (i.e., the A-T, C-G) within the genetic sequence directly informs each cell which amino acids to use when it is building a protein. Each three nucleotides (known as a codon) translate into one amino acid within the protein sequence.
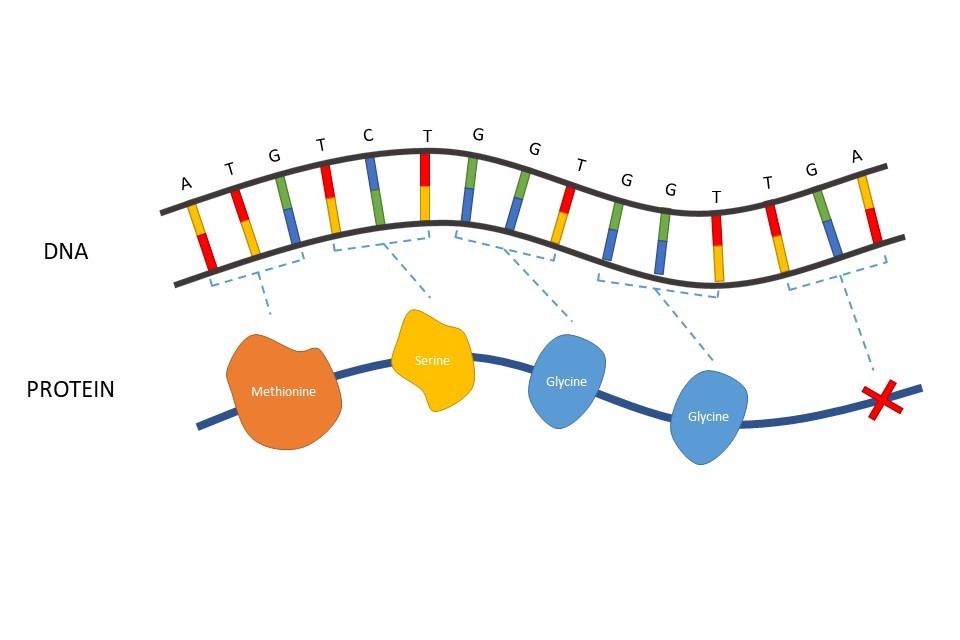
Figure 2: How each three nucleotides (a codon) in a gene can be translated to determine the amino acid order in a protein. TGA is a codon which signifies the end of a protein coding region of DNA.
For example, Figure 2 illustrates an example DNA sequence. The first codon of ATG (adenine, thymine, guanine) would produce the instructions for a single amino acid of methionine. A second codon of TCT after this would correspond to another amino acid, a serine, to be added after the methionine. The sequential addition of amino acids gradually builds a full protein. Some codons do not encode for an amino acid, but instead code for amino acid additions to stop once the protein is complete. TGA is an example of such a codon. A more detailed explanation of how codons translate to amino acids, in a process known as transcription, can be found on page 33.
A note to the reader going forward.
This report focusses on the applications and impacts of genomics beyond health. However, the science of genomics is most advanced in health and disease, so we will use examples of genetic disease to illustrate these basic principles. Those principles and their impact extend far beyond health, which we will explore further on in this report.
What is a mutation?
Genetic mutation creates a variation of a gene which may be passed down or inherited, and these variants have driven the diverse range of traits between humans, from eye colour and height to blood type and predisposition to disease. That is to say, variations of our genotype (genes) have influenced our phenotypes (measurable traits), and this same principle underlies the diversity of traits in non-human organisms too.
Mechanistically, the term mutation describes the permanent process whereby the normal nucleotide sequence in a gene is incorrectly deleted, added, copied, or swapped with other nucleotides. This can cause a change in the resulting protein for which the gene encodes, or stop the gene from producing (i.e., expressing) the protein at all.
DNA mutations occur at the nucleotide scale but can affect several million nucleotides together. Mutations occur during normal cell division as we age, or from exposure to certain environmental stimuli such as cigarette smoke or dietary factors. These mutations can lead to abnormal cell growth and the development of diseases, such as cancers.
By changing the constituent nucleotides, a mutation creates a new version of the gene. All the mutation-driven variants of the same gene are said to be alleles of each other. Not all alleles are harmful however: some alleles give rise to commonly known traits, such as the ABO blood type.
The ABO blood type is a simple example of how different alleles of the same gene create different traits. All blood types in the ABO system are determined by a single gene, known as the ABO gene. There are three alleles of this gene, known as i, IA and IB. Inheriting an allele of IA means the surface of a person’s red blood cells will be coated in antigen A, making them blood type A. Inheriting IB makes them have antigen B (type B). Inheriting both IA and IB means cells express both antigen A and B, and thus are type AB, but only inheriting i alleles means they do not express either antigen – making them type O.
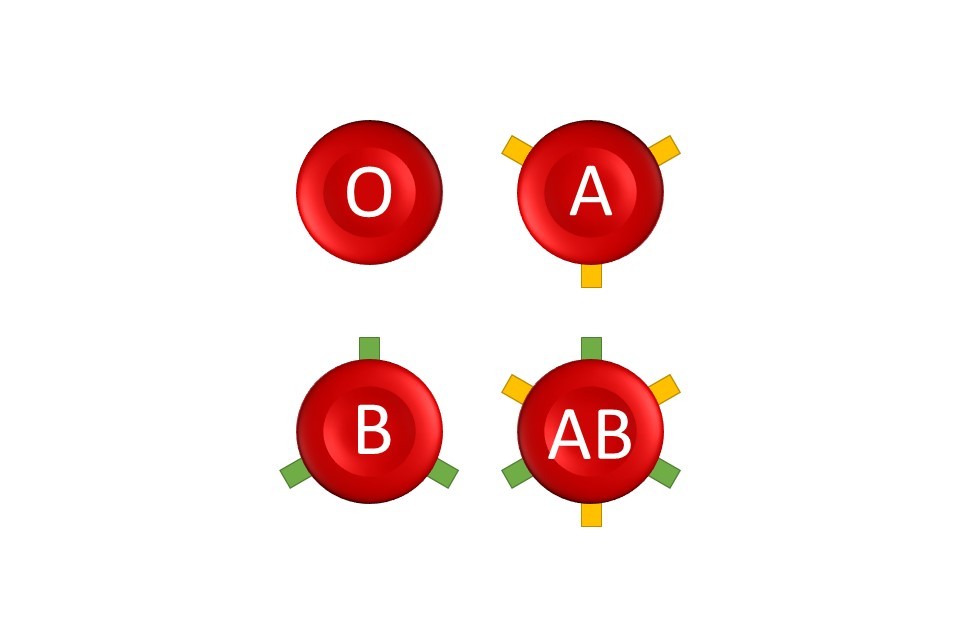
Can mutations be passed down or inherited?
Sometimes mutations can be passed between generations, but this depends on what kind of cell the mutation occurs in.
Somatic mutations are those that occur in an individual during their lifetime. An example of this is a spontaneous case of cancer which occurs with no family history. Somatic mutations only directly affect the cell in which they occur, for example a single cell in the liver. However, subsequent cell division means that changes arising from the mutation may propagate and have systemic effects in the body, such as the development of a tumour and the spread of the cancer. Somatic mutations are not passed down through families, because they do not occur in the cells which are responsible for sexual reproduction (the gametes - the sperm and the egg.)
Germline mutations are passed down/inherited through families. They are called germline mutations because they are present within an individual’s germ cells, which later give rise to their gametes. Embryos descending from a mutated gamete would also then carry the same mutation in all their cells, and this could be passed on through further generations. Germline mutations can therefore be thought of as heritable mutations.
Gene inheritance and expression
As humans we possess two copies of each gene, because each of our 23 chromosomes exist as a pair - with one copy inherited from each parent. If one of our genes is mutated on one chromosome, we have a second copy of the gene which our cells can refer to on the other chromosome, potentially reducing the effect of the mutated allele This means that the effects of the mutation might only be weakly expressed, or maybe not even expressed at all.
There are some exceptions to this however, where inheriting even just one variant gene can cause observable effects which dominate over the backup gene. This mechanism is best illustrated using two examples of common diseases, although non-disease traits often exhibit the same genetic pattern (for example, eye colour).
A trait or disease is classified as being autosomal dominant or recessive based on the inheritance pattern and subsequent expression of the gene. Huntington’s disease is referred to as an autosomal dominant disease, as a single gene mutation displays a dominant effect on the person regardless of whether they have a second healthy copy of the gene.[footnote 20]
Cystic fibrosis is known as an autosomal recessive disease, as a person is required to have two copies of the faulty gene, each one inherited from both parents. These parents may be afflicted with cystic fibrosis themselves because they carry two copies. Or they could be asymptomatic carriers for the faulty gene – meaning they themselves only have one copy of the variant and are unaffected by it. More information on the genetic causes of both conditions can be found in Figure 3.
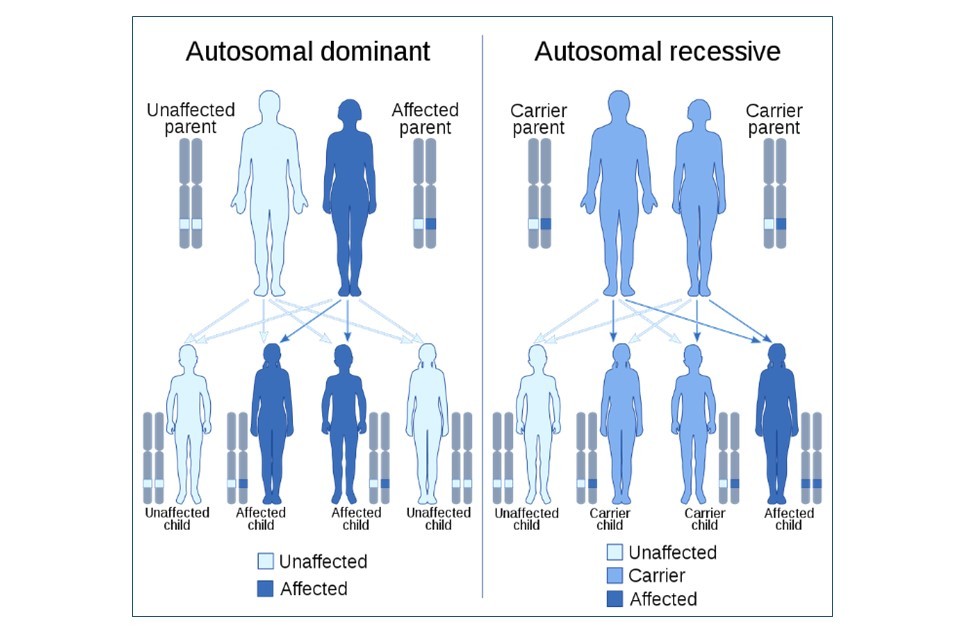
Huntington’s disease is an autosomal dominant disease caused by the inheritance of one faulty copy of the huntingtin (HTT) gene. The huntingtin protein is required for nerve function, especially in the brain. The HTT gene is approx. 200,000 nucleotides long, but in Huntington’s disease, the HTT gene is longer due to a mutation – this HTT gene contains excessive repetitions of a CAG codon, resulting in the huntingtin protein containing too many amino acids. This makes the protein too large - rendering it dysfunctional, causing irreversible neurodegeneration.
Cystic fibrosis is an autosomal recessive disease caused by the inheritance of two faulty copies of the CFTR gene from each parent, who are carriers or themselves affected. The CFTR gene is 230,000 nucleotides long, and two thirds of all cystic fibrosis cases are caused by the same deletion of a codon within this gene. This deletion causes the deletion of the 508th amino acid in the CFTR protein, which renders the final CFTR protein non-functional. Loss of the CFTR protein leaves cells unable to effectively produce sweat, digestive fluids, and mucus, causing the symptoms of cystic fibrosis.
Most of the examples above are said to be highly penetrant. This means that if someone has the gene variant known to cause a trait, it is highly likely (or even certain) that they will go on to express the trait. However, Huntington’s disease is an example where a gene can sometimes have variable penetrance, meaning that even if an individual has the mutation, the trait may develop early in life, late, or not at all. This is because the age of onset of Huntington’s disease is directly proportional to the extent of the mutation in the HTT gene which is associated with it. The age of Huntington’s onset can be predicted by determining how many repeats of a CAG codon are present within the faulty HTT gene. A person with many repetitions (equal to or greater than 60) will develop the disease at a young age, whereas somebody with fewer (36 to 40) repeats will develop it in old age. Whilst both individuals could have a faulty HTT gene, the scale of the mutation itself causes variability in the disease population.
In addition to the concepts of penetrance, gene dominance and recessivity, there are a few other ways in which genes can influence a phenotype, often by working together, or intragenomically. These are:
-
Co-dominance, as shown in the ABO blood group, is where two dominant alleles of the same gene are expressed together to make a mixed, hybrid phenotype. The AB blood group occurs when a person has both A and B alleles, which express together, making them blood type AB.
-
Incomplete dominance, which occurs when a gene exhibits a partial expression or influence in a phenotype. An example of this would be if a straight-haired person and a curly-haired person produced offspring, and the offspring had an intermediate wavy hair phenotype, which would be a hybrid of the two phenotypes.
-
Epistasis, a phenomenon whereby some genes may mask the influence of other unrelated genes. An example of this is hair colour: an individual may inherit the alleles for red hair, but if they also inherit alleles which promote baldness, the baldness may mask the influence of the red hair alleles in that person.
-
Pleiotropy, which refers to the basis that some genes may have multiple functions and affect multiple phenotypes. Disorders associated with mutations in pleiotropic genes present with a range of symptoms. For example, mutation of the TYR (tyrosinase) gene inhibits the production of melanin, leading to albinism. This is linked with the loss of pigmentation to the hair, skin, and eyes.
Are all traits caused by single genes?
Unlike monogenic (single gene) traits such as Huntington’s disease and cystic fibrosis, many phenotypic traits are caused by the combined effects of multiple genes. These are known as polygenic traits, and they often give rise to a greater range of phenotypes than monogenic traits. Examples of polygenic traits are skin pigmentation and height.
Unlike in the examples discussed previously, where one gene typically has a large effect on a trait or disease, polygenic traits do not typically follow the same patterns of dominance and recessivity. Here, multiple genes, often those of incomplete dominance, each have smaller effects and work equally with other genes to derive the final phenotype. The example of skin pigmentation/melanin production is a clear example of this phenomenon in practice.
To demonstrate polygenicity, imagine that skin pigment is caused by three genes, each with two alleles: a/A, b/B and c/C. In this example, the uppercase alleles contribute melanin (i.e., pigment) to the skin, and the lowercase alleles do not. Each person would therefore inherit six genes (three from each parent), which would combine their effects to derive pigmentation – and the specific alleles inherited of these three genes will determine the degree of pigmentation in the offspring.
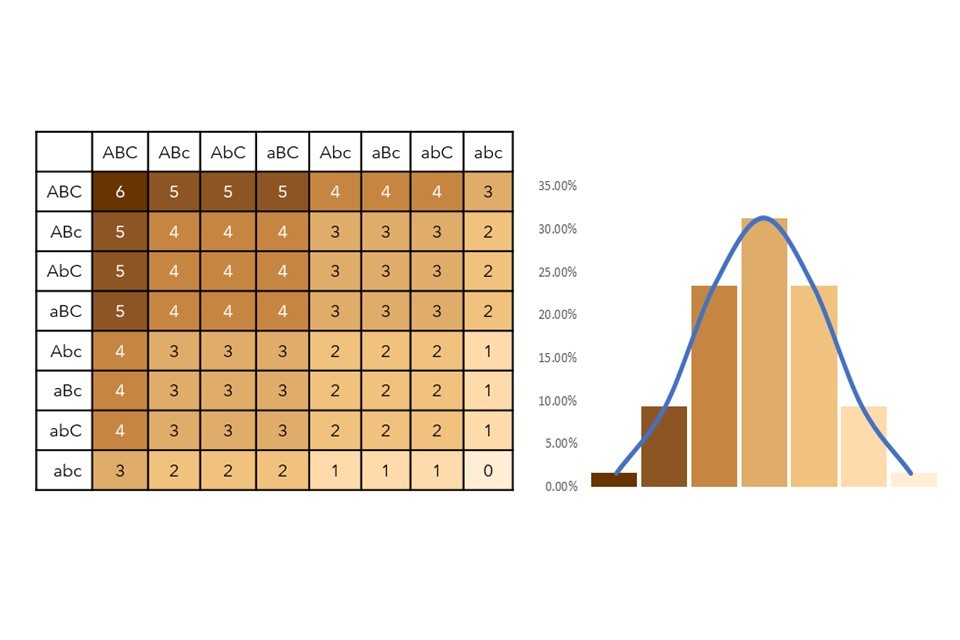
Figure 4: Polygenic scoring punnet square and frequency graph demonstrating the polygenicity of skin pigmentation.
The punnet square (numbered grid) in Figure 4 tells us all the possible pigmentation combinations in this model. It demonstrates that there are 64 genotypic combinations within this system, which would yield 7 phenotypes. For example, if two parents produced offspring which inherited ABC alleles from both parents, the offspring would be an AABBCC (ABC x ABC) genotype – this scores 6 on the punnet square and results in a dark-skinned phenotype. Offspring inheriting aAbBCC (aBC x AbC) would score 4, leading to strong-moderate pigmentation, and aabbcC (abc x abC) would score 1, a weak pigmentation. When skin pigmentation score is plotted against frequency within our hypothetical population (as shown in the bar graph), the data fits a normal distribution and a pigmentation of 3 is shown to have the highest frequency.
Polygenic traits are multifactorial, and so many are difficult to illustrate simply. For example, approximately 700 genes have already been identified to associate with height and 1,100 to associate with educational attainment, with each having a tiny effect.[footnote 21][footnote 22]
The number of genes identified to associate with traits will continue to increase as genomic studies get larger.
Epigenetic control of genes – another layer of gene regulation
As well as the key concepts of gene inheritance, alleles and polygenicity determining our traits, there is one other significant genetic factor. Epigenetics is the study of how heritable changes to a person’s phenotype (observable traits) can occur that do not involve alterations in their genotype (i.e., their genetic sequence.). Unlike gene mutation, epigenetics is a reversible process. Differences in gene expression arising through epigenetic modifications may be influenced by our individual upbringing and environment. But to discuss exactly how epigenetics works, the scale of impact it has and the importance of this in the context of this report, we must first dig deeper into how our DNA is packaged inside our cells to form chromosomes.
How is DNA packaged?
If you uncoiled all the 46 chromosomes present in just one cell and laid the DNA strands end-to-end, the DNA would stretch to be almost two metres long. The ability to package this huge length of DNA into a cell 1/100th of a millimetre wide is due to the binding and coiling of DNA into a condensed fibre, known as chromatin, and the further condensing and coiling of chromatin into the familiar X-shaped chromosomal structure.
The structure of chromatin consists of the DNA itself, which every few hundred nucleotides is wrapped around a bundle of eight proteins known as histones, much like beads on a string, to form nucleosomes. This is illustrated in Figure 5.
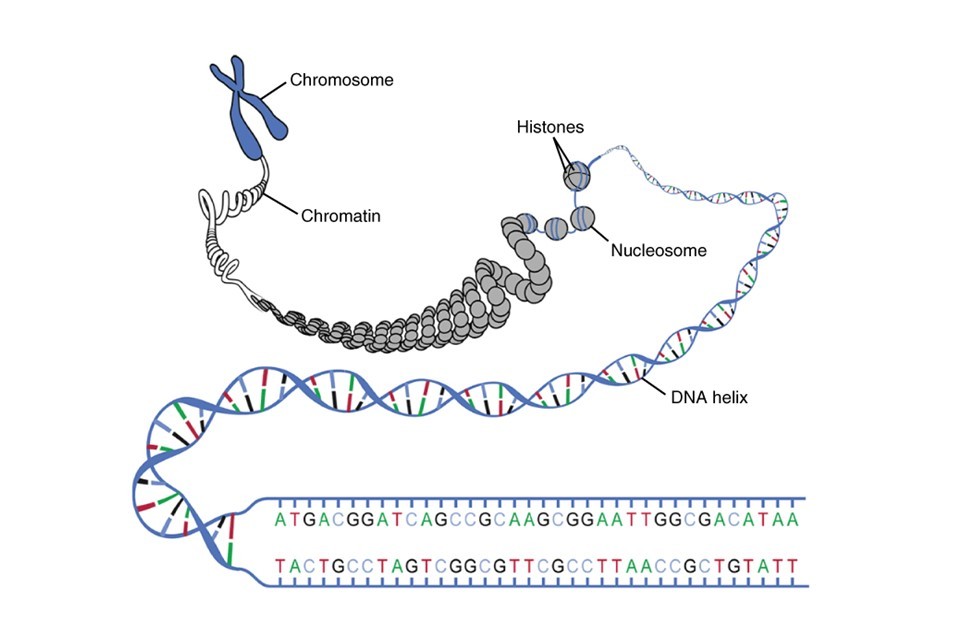
Figure 5: How supercoiled DNA forms chromosomes, and where histones are involved in this process. Image obtained from Wikimedia under Creative Commons.
But this structure isn’t just useful for packing up DNA – it can also control gene expression. Most cells in the body contain a complete, identical copy of an individual’s DNA: the long spindly cells in the brain contain the exact same DNA as the surface layer of cells in the lungs, for example. However, even though they share the same DNA, these two types of cells look different, and have completely different functions within the body. This is because cells do not express all their genes at the same time - each cell can express different genes from their DNA depending on the cellular environment they’re in.
In essence, cells can turn genes on or off depending on when or if they are needed for the cell to function properly. For example, cells in the top layer (epidermis) of skin express keratin to maintain structure and waterproofing. Cells inside bone, which have the same DNA, don’t express keratin because it’s not needed for bone to function.
To turn genes on or off in these scenarios, cells can use epigenetic modification. These modifications are reversible biochemical changes which can be applied to genes in one of two processes, but they affect DNA in similar ways.
The first way is modification of the histones which wrap up the DNA, as shown in Figure 5. These modifications either condense or relax the structure of DNA – a relaxed structure allows transcription to take place and thereby switches a gene on, whereas a condensed structure blocks transcription to switch a gene off. The second type of modification is through DNA methylation – this can also switch the gene off, similarly by coiling the DNA tighter and blocking transcription.
Transcription is part of the process of making a protein. For a gene to make a protein, the cell makes an intermediate copy of the gene known as messenger ribonucleic acid (mRNA), which can be thought of as analogous to a photocopy of a document. The mRNA is made by an enzyme known as RNA polymerase, which attaches to the beginning of genes, and reads the bases to make the mRNA copy – when the copy is made, the finished mRNA is then translated into an amino acid sequence in another part of the cell, known as the ribosome, and this is what makes the protein. However, if the DNA is tightly coiled together, such as through epigenetic modification, RNA polymerase is physically unable to bind to the gene, blocking the process and effectively turning the gene off.
The precise biochemical mechanisms by which DNA and histones can be modified to initiate or block transcription falls beyond the scope of this report. However, what is important is to appreciate is the following:
-
Epigenetic modifications are reversible – they are used to control which genes are on or off at a given time or in a given environment.
-
Epigenetic modifications are not necessarily determined by our genes, nor are they always detrimental; epigenetic regulation is a normal part of cell biology and of normal growth and development.
-
Epigenetic modifications to DNA may exert a similar scale of influence over traits as DNA sequence does.
-
Epigenetic changes, and thereby gene expression, can be influenced by external environmental factors, like childhood social deprivation , work-related stress or cigarette smoke.[footnote 23] [footnote 24] [footnote 25]
-
Understating the influence of epigenetics/epigenomics on trait development and expression may risk overstating the role of genomic influences.
From genetic to genomic science
This chapter provides an insight into how genomic technologies have progressed since the first sequencing of the human genome in 2003. The falling cost of genomic technologies is discussed, the reasons for this, and how this has changed the way genomics is employed for research purposes. The chapter concludes with a section on the limitations of current genomic technologies, and how these might be overcome with the adoption of new sequencing methods or alternative research approaches.
Key messages:
-
Recent technological improvements and reductions in cost have dramatically increased the scale of genomic sequencing.
-
Comparing a sequence to a reference genome allows you to spot the variations present (also known as genotyping).
-
Genotyping has long been used in research studies, and is increasingly offered by commercial companies for the purposes of health or ancestry analysis.
-
Studies or companies tend to use techniques that measure certain sections of a person’s genome rather than reading the entire sequence. This is known as the microarray method. Microarrays have historically been easier, quicker, and cheaper to use than full genome sequencing, but they come with some limitations.
-
Large-scale genomic studies to date have only been able to identify a fraction of the genes behind many traits we understand to be genetically driven or influenced. The inability to identify all the relevant genetic markers for a trait is known as the “missing heritability” problem.
-
As sequencing costs continue to fall and the technology continues to improve, more researchers are switching to genome sequencing rather than microarrays. Genome sequencing can overcome many of the microarray limitations, so studies built on that data are less affected by the missing heritability problem.
-
This means that we are beginning to understand more about which genes work together to influence a trait. From these observations, it is possible to calculate a predictive value for developing that trait through the genomic analysis of an individual - this is known as polygenic scoring.
-
Polygenic scores are now becoming more widely used in research, with the aim of guiding clinical intervention at an individual level. However, many polygenic scores do not predict their target trait well enough, and are therefore inferior to other traditional methods, such as a blood test. Nevertheless, as genomic data rapidly improves, so do the polygenic scores.
-
This means that in the future, polygenic scoring could provide clinical risk information far earlier than is possible with traditional diagnostic tools.
A brief history of human genomic science
Public interest in genomics increased significantly with the first publication of the entire human genome in 2003 by The Human Genome Project (HGP). The HGP was a publicly funded international collaboration launched in 1990, which the UK was heavily involved in.[footnote 26]
It aimed to establish the sequence of nucleotides which made up the entire human genome, and to determine the sections of DNA sequence that were protein-coding genes.
This was an exciting and challenging undertaking which, after 13 years, was successful in providing a fundamental understanding of the human genome.[footnote 27]
However, revolutionary as sequencing the genome was, it did not provide all the answers that researchers were looking for. They were not yet able to cross-reference complex, polygenic traits to their mutations in the genome, establish the role of the environment in gene mutation, nor could the findings provide insight into the patterns and mechanisms of genetic inheritance. Essentially, the HGP made a huge step forward in understanding what the sequence was, but this did not automatically enable understanding of what it all meant.
Following on from the HGP, several projects and consortiums were launched which sought to build upon these initial findings, allowing the field of genomics to grow substantially.
In addition to the UK programmes noted in the timeline below, additional key projects of note include:
- The International HapMap Project, which concluded in 2010, studied how clusters of alleles can be inherited together from a parent in blocks known as haplotypes, and therefore how the presence of some mutations may infer the presence of other specific mutations.[footnote 28]
The findings of the HapMap project helped us to understand how polygenic, environment-driven traits and diseases, such as diabetes and cancer, can be inherited, particularly if the genes responsible were near to each other on the same chromosome.
-
The Encyclopaedia of DNA Elements (ENCODE) project, which is currently in progress, builds upon the HGP by investigating the functions of non-coding DNA sequences, long believed to be ‘junk DNA’, and how in particular these may regulate other sections of DNA.[footnote 29]
-
Human Genome Project – Write (HGP-Write) , a follow-up project to the HGP, aims to create an artificial version of the human genome from scratch using synthetic genomics approaches, and using the results gathered from the HGP as a reference code. A sister project, GP-Write, is focussing on creating synthetic microbiological organisms, such as bacteria and yeast.[footnote 30]
-
The Darwin Tree of Life Project , led by the Wellcome Sanger Institute, aims to sequence the genomes of all 70,000 species of animals, plants, fungi, and protists resident in the British Isles, to aid conservation and to understand ecosystems.[footnote 31]
The data generated will also feed into the multinational Earth BioGenome Project, which will sequence all complex life on Earth.
A timeline of genomic science

The relatively recent explosion of genomic research is reflected in the increasing use of genomic datasets in academic publications, which now exceed those from x-ray or electron microscopy studies (Figure 6a). There are several factors behind this trend: cost (6b), speed, and the availability of reference datasets.
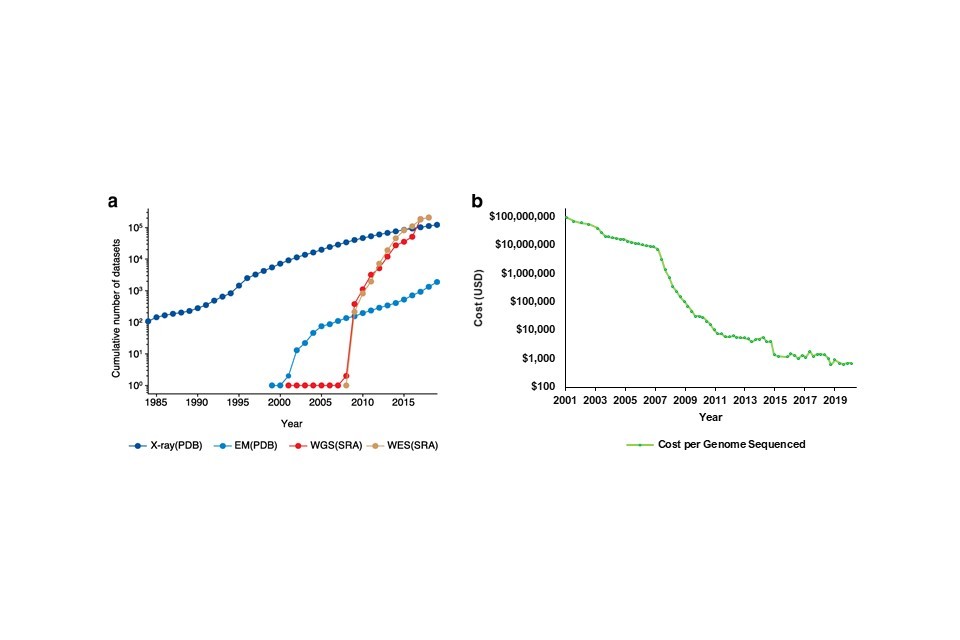
Figure 6: The increased interest, use and publication of genomic technologies is observable over time. This trend has resulted in more academic publications including genomic datasets (a), which coincides with the falling cost of sequencing technologies since 2007 (b). Key for (a): EM – Electron Microscopy, WGS – Whole Genome Sequencing, WES – Whole Exome Sequencing. Data in (a) available here, data in (b) available here.
Since 2001, the cost of sequencing a whole human genome has fallen from $100M to just under $1,0001 (Figure 6B). The precipitous reduction in cost from 2007 onwards can largely be attributed to an industry-led switch from first-generation sequencing methods that sequenced a single DNA fragment at a time (also known as Sanger sequencing) to second-generation methods that sequenced millions of fragments simultaneously. This increased the accuracy and effectiveness of sequencing technologies, resulting in a lower cost per genome sequenced. Introduced in 2005, this is known as next generation sequencing (NGS).
The increased availability of high-performance computing power has enabled the switch to NGS and meant that genomes can now be sequenced quicker than ever before; from 13 years for the first human genome to less than a day now.
There has also been an increase in the number of high-quality genome reference databases. These databases are assembled using high quality sequenced DNA from individuals which have a low number of gaps and errors. They therefore provide a guide as to the DNA sequences that occur in a species. Using these databases as a reference for comparison speeds up the process of sequencing an individual’s genome.
Next generation sequencing
Next generation sequencing (NGS) requires a sample of DNA to sequence – and there are several biological sources available which contain DNA suitable for analysis. These include:
-
Adult cells, such as white blood cells or oral mucosa cells from a saliva sample.[footnote 32]
-
Embryonic or foetal cells obtained during in vitro pre-implantation or prenatal diagnosis procedures.
-
Cell-free foetal DNA, which is present in small quantities in the blood of the mother during pregnancy.
-
Gametes, such as sperm and egg cells.
Sample Preparation
Once a sample is obtained, the DNA is isolated, and its long strands are broken into shorter fragments of about 150 nucleotides. After this step, subsequent methods can vary between manufacturers. A popular technique is described below.
Following sample processing, biochemical adaptors are then added (ligated) to the ends of the short segments of isolated DNA fragments, which allows them to be stuck down onto a solid slide or chip, as shown in . Once attached to the slide, the DNA strands are then amplified (copied) multiple times through a process (known as polymerase chain reaction (PCR) that copies the original DNA sequence: essentially ‘copy and pasting’ the DNA fragments attached. This means that the same DNA strands can now be read hundreds of times, which reduces the likelihood of random genotyping errors during the sequencing process.
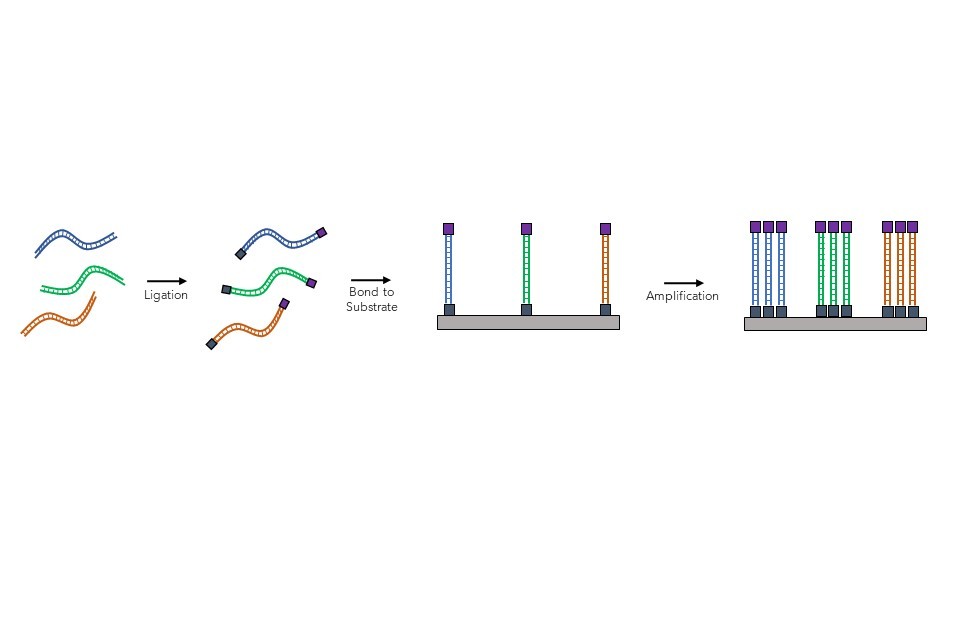
Figure 7: Sample preparation steps for NGS, involving the binding and amplification of short DNA fragments onto the substrate.
Sample Sequencing
Once the slide is fully populated with DNA strands, the strands undergo another round of amplification, except this time, fluorescently labelled nucleotides (Figure 8) are added into the reaction.
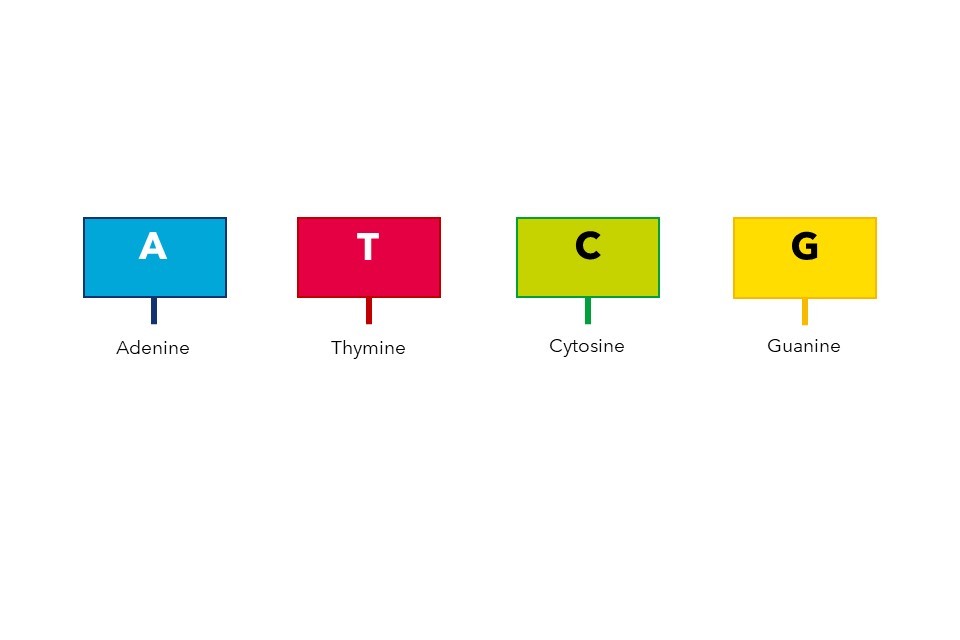
Figure 8: Nucleotide bases can be biochemically modified to fluoresce specific colours. In the following example, adenine is blue, thymine red, and so on.
When each coloured nucleotide is added to the DNA strand, the fluorescent molecule attached to the nucleotide releases a burst of light. By using cameras that can read the different colours released at each nucleotide addition, it is possible to identify which sequence of nucleotides are added to the DNA strand, which effectively indicates the genetic sequence of the DNA being assessed. This process is shown in Figure 9.
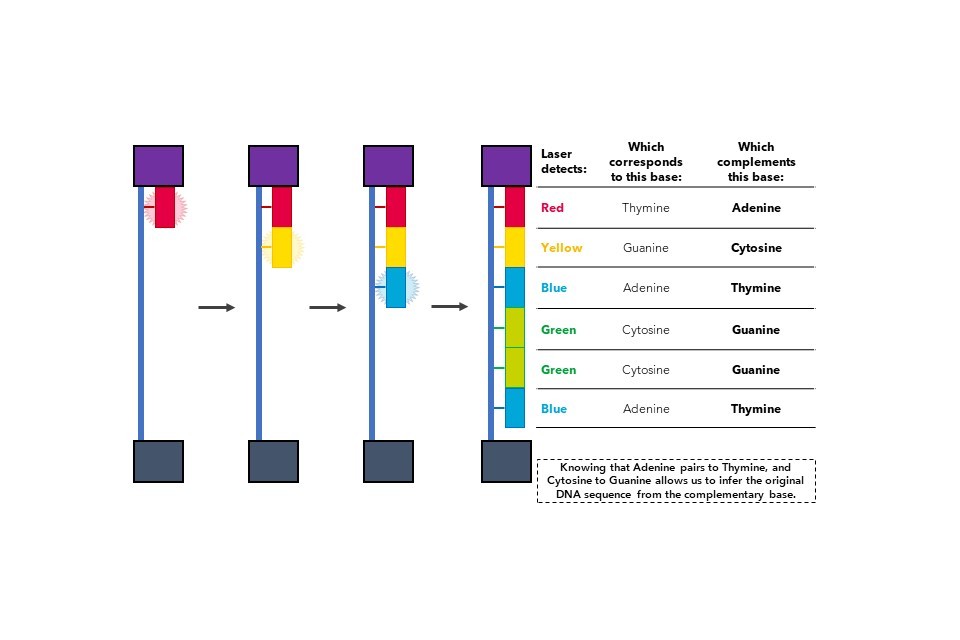
Figure 9: Fluorescent nucleotide incorporation into DNA indicates the DNA sequence during NGS.
Rebuilding the sequence
Once millions of fragmented DNA sequences have been read and the colours converted to nucleotides, the sequencing software pieces them back together into a single digital sequence by calculating where each fragment overlaps with one another. This process can be aided by cross-referencing the read-out with a standard genome, such as the genome produced by the HGP, or the software can assemble the sequence by itself if the overlaps are consistently accurate.
The advent of NGS has been perhaps the largest factor in driving down the cost of whole genomic sequencing since the mid-2000’s. NGS underpins two key sequencing approaches that are each able to sequence different amounts and sections of the genome:
-
Whole Genome Sequencing (WGS) involves sequencing the entire (or near entire) DNA within the genome, including both coding and non-coding DNA regions.
-
Whole Exome Sequencing (WES) involves only sequencing the protein-coding portion of DNA within the genome, leaving the non-coding regions of DNA unsequenced. However, a growing body of research suggests that non-coding DNA might be just as responsible for many mutations or traits, which this method would fail to identify.
What’s next for sequencing technology?
Despite the impact of NGS on the field, new ways of sequencing are being developed, known as third generation sequencing technologies. These can perform longer sequencing reads, meaning that DNA requires less processing prior to sequencing. Some third-generation sequencing technologies are already commercially viable and used in research, including some that are portable. Some of these technologies have higher error rates than older technologies, but improvements in this area are developing rapidly.
Third generation technologies can simultaneously assess genetic sequences and epigenetic modification on the same read. They also have the potential to sequence other biomolecules, such as DNA-protein intermediaries (mRNA). Together, this may mean that the advent of third gen methods could very well prove as revolutionary to the field as NGS was back in 2007.
The UK has maintained a world leading position in the development of sequencing technologies. First generation (Sanger), second generation (Solexa), and leading third generation (Oxford Nanopore) technologies were all developed in the UK.[footnote 33][footnote 34][footnote 35][footnote 36][footnote 37]
NGS vs. DNA Microarrays
Aside from sequencing someone’s DNA, there is an alternative method to determine someone’s genotype, but this method comes with a few more limitations. The method is called DNA microarray, and it was first developed during the 1980s as a way to obtain genomic data from participants cheaply, quickly, and easily. This is because although Sanger sequencing was possible then, it was slow and expensive, and NGS wasn’t yet invented. This meant that DNA microarray was a good middle ground to facilitate research.
DNA microarrays are essentially small, commercially available chips which contain probes complementary to specific DNA sequences. When a sequence of DNA corresponds to the probe, they bind together and emit a signal, indicating that the DNA sample contains that known sequence of DNA. So, whilst the microarray does not actually sequence the nucleotides of DNA, it can find out if a known DNA variation is present by applying a probe for that sequence.
The results, or ‘hits’, from the probes can then be applied to predict genomic variation in areas of the genome not directly assessed by the probes, in a process known as imputation. Imputation takes the hits obtained by the microarray and compares these to hits found on reference genomes (i.e., in other people). It can then predict the presence of gene variants in other areas of the individual’s wider genome based on probability, by comparing these hits to many others, without genotyping those variants directly.
DNA microarrays can therefore allow you to survey an individual’s DNA for known gene variants of interest, and then to make some conclusions as to their wider genome through imputation. This approach differs from sequencing, which determines the sequence of DNA nucleotides from scratch. Some additional pros and cons to microarrays are considered in Table 1.
Table 1: The pros and cons of using microarrays to obtain genomic data.
| Pros | Cons |
|---|---|
| Microarrays are easy to use, with well adopted/established protocols | Detection of the genes is dependent on the quality of supplementary technology (chip readers etc) |
| Chips are available for lots of different genes and alleles | Cannot detect unknown gene alleles, so may introduce bias by missing subpopulations |
| Simple data outputs – does not require high computing power for analysis | Potential errors if DNA probes bind non-specifically to random DNA, and give false positives |
| Currently (and historically) cheaper to use | Price saving could be overtaken by NGS in the next few years |
Microarrays have been the predominant method for obtaining genomic data until now because a key consideration in research is cost. This means that the bulk of studies built on that data indirectly rely on microarrays. However, the limitations of microarrays outlined in Table 1 have shaped the reliability of that data, which in turn can limit the power of the studies which make use of it. We will discuss the effects of those limitations in Chapters 4 and 5.
Using sequencing to determine genotype and phenotype
Genomic knowledge, particularly before the publication of the human genome and the development of NGS and DNA microarray, was developed heavily from twin studies. By observing differences between identical (monozygotic, sharing 100% of DNA) and non-identical (dizygotic, sharing ~50% of DNA) twins during childhood and adulthood, scientists could estimate the extent to which traits may be influenced by genomic and environmental factors.
Twin studies led to the development of heritability estimates. These are values assigned to traits based on how much of the variance in a trait is statistically estimated to arise from genotypic similarity (compared to the amount estimated to arise from environmental factors and random error). For example, if a trait had a heritability of 0.80, it would indicate that 80% of the variation in the trait can be statistically explained by DNA differences between individuals in the twin sample. Conversely, 20% of trait variation could be explained by differences in environments or statistical error. Table 2 outlines heritability estimates for several traits obtained from twin studies.
Table 2: Heritability estimates for a broad range of traits.
| Trait | Heritability | Study or population | Reference |
|---|---|---|---|
| Acne | 85 percent | Twin study (Australian participants) | [footnote 38] |
| Autism spectrum condition | Up to 95 percent | Twin study (UK participants) | [footnote 39] |
| Body mass index (BMI) | 47 to 90 percent (age variable) | Meta-analysis of 88 study estimates (mixed population) | [footnote 40] |
| Major depressive disorder | 38 percent | Twin study (Swedish participants) | [footnote 41] |
| Hair colour | Up to 99 percent | Twin study (Dutch participants) | [footnote 42] |
| Handedness (left versus right) | 25 percent | Twin study (Australian and Dutch participants) | [footnote 43] |
| Intelligence | 20 to 80 percent (age variable) | Twin study review (diverse samples) | [footnote 44][footnote 45] |
| Musical ability | 21 to 51 percent (test dependent) | Mixed populations | [footnote 46] |
| Schizophrenia | 81% | Meta-analysis of 12 twin studies (diverse population) | [footnote 47] |
Heritability estimates are expected to vary across samples, even where genomic influences are exactly consistent. This is because environmental differences exist over places and time, and their effects on people vary between contexts. Differences in trait variation due to environmental and residual factors between samples is therefore expected to lead to different heritability estimates.
An example of this is the heritability of intelligence, which ranges from 20 percent to 80 percent depending on the age of the sample analysed, increasing steadily as people age44,45. This is thought to be due to changing environmental influences throughout life, though these could in turn be part driven by genomic differences whereby people select into specific environments depending on their genome. This is referred to as gene-environment correlation (rGE).[footnote 48]
While heritability estimates can vary according to context, they are still used as a broad measure for indicating the role of genomics in the formation of traits in a particular population at a particular time.
Applying heritability to genetic science
The development of heritability estimates was an important step in learning about gene-trait interactions, because it meant that researchers (using twin studies) could estimate what percentage of a trait was genetically influenced, without having to establish which specific genes were behind the trait.
Then, as genotyping and computing power advanced rapidly in the mid-to-late 2000’s, it became possible to carry out genomic studies en masse, far beyond the scale of previous studies. The falling cost and growing availability of genomic technologies facilitated the collation of large-scale genomic datasets across large populations, which was further supported by international genomic collaborations, biobanks, and genome projects.
This led to the development of genome-wide association studies (GWAS). These were proposed as a method to scan the entire genome to see which genetic mutations or variants are associated with traits of interest. GWAS were heralded as the counterpart solution to twin study-derived heritability estimates, as they could be used to identify specific alleles and variants behind high heritability traits, which, until now, had remained unidentified.[footnote 49]
However, GWAS could not determine which mutations caused a trait, only those that were statistically associated with it. Whilst offering many clues about how traits came to be developed and which gene variants were involved, in many cases GWAS couldn’t conclusively identify causal genes. Nevertheless, the use of GWAS techniques has led to the successful identification of gene variants involved in several traits , thereby advancing and improving the treatment of a variety of conditions such as age-related macular degeneration, prostate cancer, and type 2 diabetes.
What is a GWAS and how does it work?
Essentially, a GWAS takes large-scale genomic datasets and looks at whether each genetic variant is associated with a trait. In the case of binary traits (for example developing a disease versus remaining healthy), a GWAS examines whether a variant is more frequent in a trait group when compared to a non-trait control group. In the case of continuous traits, such as blood pressure, a GWAS examines whether a variant is more common amongst those with higher or lower values of the trait. Because genotype is set at conception and (generally) cannot be influenced by the environment, associations between a gene variant and a trait infers a potential causal role of the variant.
The first stage of GWAS is the collection and collation of genomic data, which is later computationally mined and analysed for associations. There are two main methods to collect DNA data, NGS and DNA microarray, which were discussed earlier on pages 40-44. The most common approach to data collection has been through the DNA microarray technique. The NHGRI-EBI Catalogue of human genome-wide association studies lists 5,059 GWAS based on microarray data, compared to only 358 sequencing-based association studies.[footnote 50]
This has been due to the prohibitive cost and duration of whole exome/genome sequencing at the time. However, the microarray approach does have several limitations, and this in turn has historically limited the predictive power of GWAS.
The primary challenge with the DNA microarray method is that you first need to know which genes and alleles to survey for a trait before commencing any analysis. This contrasts the whole genome sequencing approach of sequencing every nucleotide. This pre-selection step can introduce bias into GWAS design, as variant genes of interest may only be present in the specific group originally studied, such as those of European descent or men. This could mean you might fail to capture other variants present in other populations, even if you were to include them in the study.
Another limitation is that microarrays tend to look only at one specific type of mutation called a single nucleotide polymorphism (SNP). A SNP is defined by one single nucleotide (A, T, C or G) being swapped for another nucleotide within the germline genetic sequence. Whilst SNPs are the most frequently occurring type of mutation in the human genome, with a person having on average 4-5 million SNPs within their genetic code, mutations other than SNPs (such as insertions, deletions or codon duplications, shown in Figure 10) can have just as much influence on gene function as SNPs do. Therefore, inadvertently only probing for SNPs may overlook non-SNP gene variants.
Figure 10: Variations in the types of DNA mutation can have different levels of downstream effect.
A sequence of ‘GCC TAC CGA TCG’ translates to the amino acid (AA) sequence ‘Ala Tyr Arg Ser’. Some SNPs affecting this sequence may be silent (aka synonymous) – as both GCC and GCT code for alanine, this SNP will lend no recognisable effect to the protein. However, a one nucleotide change from GCC to GTC changes Ala (alanine) to Val (valine). Similarly, a change from TAC to TAA causes the AA sequence to end prematurely (a nonsense mutation), which deletes the rest of the protein. There are other kinds of mutations in addition to SNPs, and these can often have more of an effect. This is because they can cause a frameshift, which moves the entire downstream triplicate sequence out of sync. Examples of these are insertions, deletions, and duplications.
DNA microarrays are also limited by the fact that, unless specifically adapted, they only investigate the basic genetic sequence within a genome. This means that they can ignore any epigenetic modifications to the genes present which, as discussed earlier, may be as important to a trait as the sequence of nucleotides. They also do not provide any insight into potential complex gene interactions behind polygenic traits, only the extent to which each SNP is associated with the trait.
The limitations of DNA microarrays have meant that whilst GWAS have been a powerful tool, they have only investigated certain parts of the genome. This means that when they have been used to explore the genetic basis of high heritability traits, GWAS have only been able to identify a fraction of the genes which are associated with it, partly for this reason.
An example of this in practice is the study of height. A twin study from 2003 estimated the genetic heritability of height to be between 73 to 81 percent in European ancestry populations.[footnote 51]
However, a subsequent GWAS performed using DNA microarray data in 2010 (on an Australian population), could only predict approximately 45 percent of variation in an individual’s height, leaving an unexplained genetic gap of around 32 percent.[footnote 52]
The discrepancy between the heritability of traits estimated from twin studies and the genes that GWAS have identified has been termed the missing heritability problem.[footnote 2] The missing heritability problem arose because GWAS studies are statistically underpowered to reliably estimate the small genetic associations for all variants in the genome. Furthermore, they have commonly excluded uncommon and rare variants, and are unable to model the presence of non-additive effects (gene to gene interactions) or the presence of gene-environment interactions.
Furthermore, a GWAS can be poor for predicting some traits, especially if multiple genes are involved in the trait (such as the trait is polygenic). This is because many polygenic traits are influenced by genes that each have a small effect, contributing accumulatively to the trait. In addition, mild genetic influences on a trait may be easily masked or overruled by the expression of other genes (see intragenomic influences in Chapter 2) or be modified by environmental exposures through epigenetic mechanisms.
Finally, GWAS have thus far generally been conducted on very limited samples. In addition to the bias introduced by microarray limitations, recruitment for the study cohorts is also highly non-representative of populations (as shown in Figure 11). Research has shown that as much as 83% of GWAS were conducted on samples of only European ancestry participants.[footnote 4] This is problematic for several reasons. First, Individuals of European ancestry represent only a portion of the global community and therefore Eurocentric ancestry studies will only capture a small amount of the total human genetic variation. Second, populations differ, meaning that results are unlikely to be applicable across ancestries. Genomic prediction may therefore be highly inaccurate across diverse populations, reducing the effectiveness of genomic applications to all citizens.[footnote 53][footnote 54]
Third, it may be difficult to remove the effects that social stratification and geography have on traits.[footnote 4][footnote 55] Finally, it may lead to reduced engagement in research from marginalised groups.
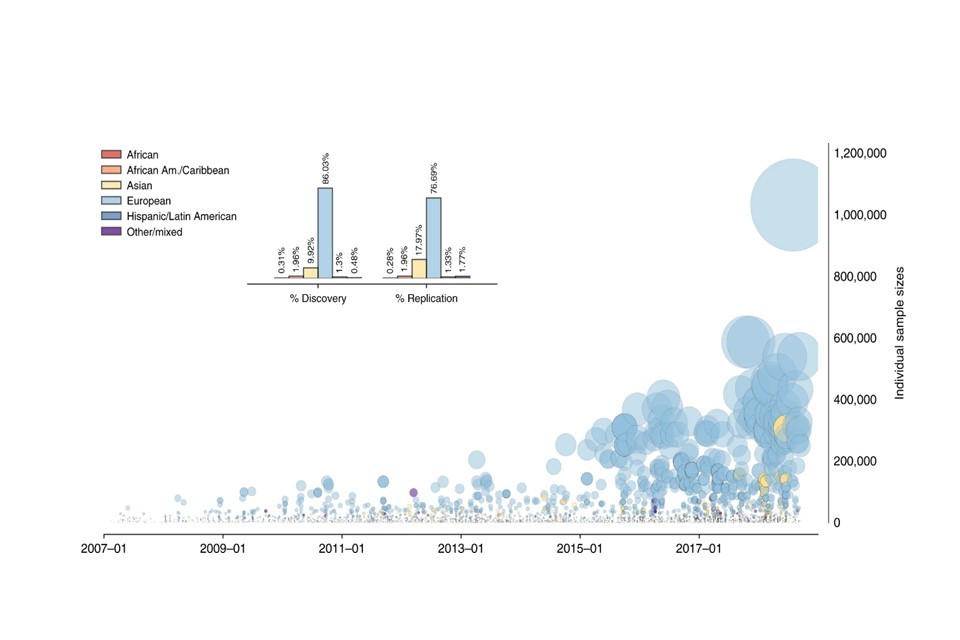
Figure 11: GWAS cohort sizes are growing - but they are still largely based on populations of European ancestry.[footnote 4] Cohort sizes of GWAS have been growing since 2007, but these increases are largely focused on data obtained from those of European ancestry. Over this period, 86% of discovery studies and 77% of replication studies were based on European data. Those of African ancestry have the lowest representation, at 0.31% and 0.28% respectively. This raises questions as to whether GWAS findings would be as applicable in understudied populations.
Despite these drawbacks, microarray driven GWAS studies are the main source of information for many traits, providing relevant genomic information that has potential use for predicting trait risk and identifying potential causal mechanisms.
However, future applications of GWAS are likely to turn towards a more whole genome sequencing approach as a way of minimising several of the above limitations and some of the sources of bias within GWAS study design.[footnote 56]
It is thought that this change in approach will improve the reliability and replicability of GWAS when identifying trait-associated genetic variations.
Some early studies are now demonstrating that this is the case– including for height variability, as discussed earlier. These studies have shown that whole genome sequencing approaches for determining height can identify approximately 79% of variation between individuals , falling within the original heritability estimate of 73-81 percent.[footnote 57]
This is because sequencing approaches are better able to identify rare, low frequency gene variants when compared to microarray/imputation methods.[footnote 58]
Therefore, as genome sequencing becomes more prevalent, GWAS performed using data obtained by that approach will become more robust and reliable, and hereby the findings will be more relevant to decision making processes in the future
Polygenic scoring as a predictive tool
Despite the current limitations of GWAS, the findings and outputs of such studies are still informing the development of diagnostic genomic tools. While individual SNP-trait associations are commonly small and offer very little prognostic power, combining all observed SNP-trait associations in an additive fashion can provide a powerful tool. This approach, referred to as polygenic scoring, calculates a weighted-sum of an individual’s combined trait-associated alleles (as identified by GWAS). These polygenic scores may then be further combined with other relevant characteristics, such as lifestyle factors, to derive a more accurate overall predictive value for a trait.
Polygenic scores may be used within medical contexts to identify individuals at high risk of disease for early intervention. A breast cancer polygenic score, in conjunction with known clinical risk factors, has been found to identify 16% of the population who could make an informed decision to start breast screening at the earlier age of 40 years of age instead of at the recommended age of 50.[footnote 59]
The same process can identify those at sufficiently low risk and who could therefore delay screening. Personalisation is a familiar concept in medicine and the use of genomic information to extend this personalisation has the potential to improve treatment for patients.
Polygenic scoring approaches can also be applied to non-medical traits. This could be for example with the aim of establishing more effective ways for teaching children and improving their educational development. Interest in polygenic scoring in fields beyond health is growing as the genomic evidence base develops, though the scientific base for many non-health traits is currently less well developed than for some health traits.
It is crucial to emphasise that the predictive power of a polygenic score is dependent on several factors. Polygenic scores are not universally accurate across all human traits, but rather vary because of several biological and data driven factors. Genomic factors include the overall heritability of the trait in question, the number of genes that influence the trait (the trait polygenicity), and the effect sizes of the genes involved (the effect size distribution). Data driven factors include the sample size of the original trait GWAS used in polygenic scoring, the frequency of the variants in the GWAS sample population, and the ancestral similarity of the GWAS sample and the sample within which polygenic prediction is being applied.
Limitations to genomic prediction
Predictions from current polygenic scores are generally less informative compared to non-genomic information that is already available. For example, an electrocardiogram (ECG) will provide more information with regards to a heart condition than a polygenic score for heart disease.[footnote 60]
Similarly, a polygenic score for predicting educational attainment (currently one of the best performing polygenic scores available) is less informative than a simple measure of parental education5. The predictive power of polygenic scores will inevitably increase with larger studies and studies based on whole genome sequencing data.
Individual level prediction is notoriously difficult for complex traits, with or without genomic information. Different challenges exist in the analysis of group differences (why do group A have poorer health than group B) and individual differences (why does one individual from group A have poorer health than another individual from group A). What appear as chance or random events can be important for determining an individual’s traits but are generally averaged out at the group level. This is particularly relevant for complex traits that are influenced by a plethora of different genomic, social, cultural, and environmental factors. GWAS results only represent averages within the context of these other factors. That is, a SNP effect estimated from a GWAS represents the average difference in a trait that is observed amongst the population of sampled individuals who carry a specific copy of that SNP. In a GWAS of height, a SNP effect would represent the average amount taller that people with a certain copy of the SNP are compared to people who had a different copy. When combined into a polygenic score, these average SNP predictions may be generally predictive of height, but the predictive accuracy may vary across individuals, who are individually unique rather than average.
Things seen in genomic data may not be ‘genomic’
Because genotype is fixed at conception and cannot be influenced by environmental factors (although genomic effects themselves can be), it is intuitive to assume that gene effect estimates in GWAS are causal. However, there are other factors that can underlie these estimates.
First, genomic differences can exist between populations at global and local scales. These differences may arise because of several factors. Spatial isolation and a lack of population mixing can make populations distinct from one another. There may be migration and founder effects, whereby the genomic variation of a population is limited to the small group who migrated to a new area. Spatial isolation can also lead to systematically different changes in allele frequencies between populations, referred to as genetic drift. Non-random mating and the selection of partners based upon certain characteristics such as height or hair colour can make populations different over time. These genomic differences are referred to as population structure and can be strongly geographically patterned; natural boundaries such as mountains and oceans have historically been barriers to population mixing.
Because human traits can be geographically patterned for social and cultural reasons, population structure can lead to ‘false’ associations between genes and traits. This is referred to as population stratification . Because of population stratfication, many GWAS are conducted on single (often European) ancestry samples.[footnote 61]
Statistical techniques are further used to minimise population stratification, but these are often imperfect.[footnote 62][footnote 63]
In addition to inheriting their genes from parents, most offspring are also raised in an environment that has been curated by their parents (the exceptions being children who are raised away from their biological parents). These curated environments may themselves be linked to parental genotype. For example, parents with a strong genetic predisposition to curiosity and learning may furnish their household with educational books. In addition to the potential inheritance of a genomic predisposition for learning, children of such parents will also benefit from being raised in an environment that is conducive to learning. Because this environment is partly influenced by the genes that children themselves inherit, genetic effects can be considered to operate both “directly” (through inheritance) and “indirectly” (through environment). These indirect genetic effects are sometimes referred to as genetic nurture and can sometimes be detectable in the genome.[footnote 64] [footnote 65]
Genomic prediction in policy and practice
Because polygenic scores cannot yet predict many traits with sufficient accuracy and have limited generalisability across populations, polygenic scoring is not yet used in any area of policy. However, the use of polygenic scores is now starting to be trialled in some areas of healthcare.[footnote 66]
As the genomic evidence base increases and more is known about the role those specific genetic markers which play in the development of traits, polygenic scores may become more widely adopted. Due to the historical scientific focus on genomics within health, advances in genomic prediction are likely to occur sooner and more rapidly within health fields than in fields beyond health. However, the increasing interest in genomic prediction beyond health and the application of knowledge developed within health genomics means that genomic applications may occur across many areas of society.
Genomics: where we are now
The greatest advances from genomic technologies are currently found within the healthcare sector. However, there are other contexts in which genomic technologies are already having an impact upon humans and their environment. In this chapter we explore five sectors in which genomics is already being applied, or applications are being trialled: genomic medicine, direct-to-consumer (DTC) testing, forensic science, synthetic biology, agriculture and food, and ecology.
Key messages:
-
The UK has an ambitious genomic healthcare strategy, Genome UK, which will lead to a massive increase in the number of human genomes available for research. This will likely allow us to unlock genomic research and applications in health and non-health areas.
-
The UK is a world leader in the development of genomic data and infrastructure, as evidenced by its position at the forefront of genomic research into COVID-19. The UK can lead internationally on ethics and regulation if we can overcome the major data and research challenges faced.
-
The direct-to-consumer genotyping market is growing rapidly. Tens of millions of consumers across the world have submitted their genomic data for commercial testing to gain insights into their health, ancestry, and genetic predisposition to traits.
-
The accuracy of genomic-based insights which underpin direct-to-consumer services can be very low. They may also impact an individual’s right to privacy, through data sharing, identifiability of relatives, and because of lapses in cybersecurity protocols. Customers may not fully understand these issues.
-
Direct-to-consumer genomic tests for non-medical traits are largely unregulated. They fall beyond the scope of current (i.e., medical) legislation, relying instead on voluntary self-regulation by test providers. Furthermore, many of these companies are based outside of the UK and are therefore beyond UK regulations.
-
Genomic sequencing may offer new opportunities within forensic science, synthetic biology, agriculture and food, and ecology.
-
Concerns exist around data consent, privacy, and security. There is currently no legislation which completely bars law enforcement from using genomic data obtained from sources other than criminal databases.
Considerations for policymakers:
- Sufficient communication between direct-to-consumer genomics companies and customers. Transparent, comprehensible information will be increasingly important, particularly when communicating:
- to what extent tested genomic markers can accurately predict each trait,
- the potential for identifying individuals (and those beyond the consumer) from their genomic data,
- the inherent risks of consumers uploading their raw genomic data to third party databases, especially from the perspectives of data privacy and security,
-
the likelihood of any future potential to predict additional traits (such as those beyond health) from existing genomic data.
-
Regulatory status of non-medical genomic tests (such as for ancestry). Results from these tests may still impart some degree of medical information to the consumer. There is therefore a question as to whether some medical diagnostic test regulations ought to apply in these cases.
-
Voluntary self-regulation amongst DTC companies. Should government not seek to impose direct legislation in this area, encouragement of further self-regulation may be advantageous to uphold market standards.
-
Law enforcement access to genomic databases. A wider discussion on the acceptability of allowing law enforcement access to genomic databases would be welcome. This includes access to healthcare/research, direct-to-consumer, or third-party databases, and could explore which databases/resources law enforcement might have access to, and the extent of that access. It would be useful to reflect on and clarify whether any of these resources would fall within scope of the Crime Overseas Production Orders (COPO) Act, which gives UK authorities the power to obtain electronic data directly from service providers outside the UK. The introduction of the COPO Act alongside international acts such as the US Clarifying Lawful Overseas Use of Data (CLOUD) could make medical data accessible to international law enforcement agencies.
- International regulation. There may be a future need for international regulation around the use and introduction of genomic editing to manage the potential risks surrounding environmental and ecological applications. An exploration of the need for such regulation and mechanisms for development would be welcome.
Genomic medicine
-
Personalised medicine
-
Rare disease research
-
Population genomics
-
Genomics and COVID-19
Direct-to-consumer testing
-
Ancestry and genealogy tracing
-
Disease risk prediction
-
Online genomic databases
-
Data protection
Forensic science
-
Enhancing forensic DNA databases
-
Missing person or cold case identification
-
Phenotypic prediction
-
Individual right to privacy
Synthetic biology
-
Technology applications
-
Genome editing
-
Synthetic genomics
-
Challenges and ethics
Agriculture and food
-
Selective breeding and genomic selection
-
Genetically modified organisms
-
Genomics for food quality and composition
-
Food traceability and provenance
Environment and ecology
-
Invasive or protected species identification
-
Ecological monitoring and contamination
-
Genomics-informed conservation
-
Modelling species adaptation
Genomic medicine
The largest human impacts of genomic technologies are currently seen within the healthcare sector. Aside from the genomic response to COVID-19, the medical applications of genomics will likely represent the first exposure to personal genomic sciences for many of the general public, thereby shaping public attitudes towards the genomic sciences more generally. The healthcare applications of genomics are also likely to greatly influence the development of national genomic infrastructure. This infrastructure will allow other genomic applications beyond health to develop. The COVID-19 pandemic has highlighted the strengths of the UK’s genomic infrastructure; it has been at the forefront of identifying and monitoring SARS-CoV-2 transmission internationally.
The history of genomic medicine in the UK
The UK Life Sciences Strategy published in December 2011, affirmed the first national commitment to integrating genomic technologies within healthcare.[footnote 67]
In late 2012, then UK Prime Minister David Cameron announced the launch of the 100,000 Genomes Project, which aimed to sequence the entire genomes of 100,000 NHS patients.[footnote 68]
The project focused on patients with cancer and rare diseases, to provide participants with targeted interventions in their healthcare.
In 2013 the Department of Health created Genomics England to oversee the project.[footnote 69]
Participants were recruited from thirteen NHS Genomic Medicine Centres across England. The first diagnoses were received in 2015 and by December 2018 all 100,000 genomes had been sequenced.[footnote 70]
One in four participants received a genetic diagnosis, bringing the prospect of genomics-informed personalised medical interventions to thousands of people.
Genomic medicine today
Following the UK Chief Medical Officer’s Generation Genome report in 2016[footnote 71], the NHS Genomic Medicine Service (GMS) was launched in 2019 to build upon the success and infrastructure of the 100,000 Genomes Project.
The GMS aims to fully integrate genomic medicine into routine clinical care, and to sequence 500,000 whole genomes by 2024. This will include all children with cancer, those seriously ill with rare genetic disorders, and adults with hard-to-treat cancers.
Outlining the wider national genomics strategy in more detail, the UK government published Genome UK in September 2020 as a 10-year healthcare strategy. It also plans the implementation of genomic healthcare via the GMS, outlining the wider role that genomic science may offer for clinical diagnosis, stratification of medical care and personalised medical treatment. There are also a number of research cohorts in the UK with large genome sequencing programmes, which will complement the work of the strategy:
-
UK Biobank: A UK Research and Innovation (UKRI), Wellcome Trust, charity and industry led biomedical research resource project with 500,000 participants, all of which will have their whole genome or exome sequenced by the early 2022[footnote 72].
-
Our Future Health (formerly known as the Accelerating Detection of Disease challenge): A UKRI and industry funded project which will be the UK’s largest ever health research programme, recruiting up to 5 million diverse participants. It aims to evaluate and provide polygenic risk scores for at least three million participants[footnote 73].
-
National Institute for Health Research (NIHR) BioResource: A Department of Health and Social Care (DHSC) funded agency, providing whole genomic sequences or genotypes of 150,000 volunteer participants[footnote 74].
What are the UK’s goals for genomic medicine in health?
To deliver the aims of the Genome UK strategy, the potential to use genomics to identify effective drugs or treatments for individuals based on their genomic sequence (known as pharmacogenomics) will be explored. Genomic data will also be combined with existing clinical data such as tumour imaging information to improve cancer treatment and research. Genomic sequencing will be applied to infectious diseases to monitor outbreaks and better identify high risk individuals, whilst whole genomic sequencing will also be used to speed up and improve the diagnostic process for thousands of individuals with inherited rare diseases[footnote 75].
Genomic data may also be used in a preventative healthcare context, building upon that already used with some forms of inherited cancer risks. This includes non-invasive prenatal testing, evaluating the potential of genomic screening for newborns, and a wider targeted genomic screening approach adopted by the NHS. The UK government has committed to exploring the ethics and risks of such preventative healthcare measures and any necessary regulation in this area. Furthermore, Genomics England and the UK National Screening Committee have recently undertaken public dialogue on the use of genomics for newborn screening[footnote 76].
The accumulation of whole genome data collected will also help to support both translational and cross-disciplinary research (research that has direct relevance to health or non-health applications). The large databases which will hold the genomic data will therefore provide a valuable tool for more informative research into health and non-health applications. The rich information of these databases may be used to supplement or even supersede non-genomic research methods and provide better predictions of traits within and beyond health.
In recognition of this potential, Genome UK has committed to ensuring all our genomic data systems will continue to apply consistently high standards for data security, while allowing appropriate use and analysis to boost health and research. It will coordinate the development of standardised data access processes, ensure the diversity and equity of data, and uphold clear standards on responsible data use.
How can genomics complement other emerging technologies?
To achieve the ambitions set out in the Genome UK strategy, the UK will rely heavily on bioinformatics, the use of computational tools to derive complex analyses and conclusions from biological data. This brings an increased need for enhanced computing capabilities[footnote 77], complementing development in high-performance computing infrastructure and the development of machine learning (ML) and artificial intelligence (AI).
How does the UK’s genomics strategy compare internationally?
Few countries are seeking to implement genomic medicine within their healthcare systems as extensively as the UK. This makes drawing direct comparisons with other nations difficult. However, it is possible to compare the UK’s past genomic sequencing project, the 100,000 Genomes Project, to other ongoing international sequencing projects.
As of 2021, there are at least 41 active national genomics projects worldwide and several more international collaborations such as the H3Africa Project (Figure 12)[footnote 78]. Further details of these can be found in the Appendix. The main aims of national genomics projects tend to be similar, and include:
-
Establishing databases of normal genomic variants (an aim of 90 per cent of projects, globally)
-
Determining pathological gene variants, such as those involved in rare diseases (71 per cent)
-
Building the infrastructure to support wide-scale genomic sciences (59 per cent)
-
Implementation of personalised medicine into clinical care (37 per cent as a direct benefit of the program, but as a rationale behind 85 per cent of projects.)
Sequencing targets and levels of investment for national programmes vary widely The smallest programme aims to collect 80 sequences (The Uruguay Urugenome project) while the largest aims to collect 100,000,000 sequences (the Chinese Precision Medicine Initiative). The Chinese Precision Medicine Initiative receives the most funding (a total of CNY ¥59.8 billion or GBP £6.6 billion), whilst Slovenia’s Genome Project receives the least (GBP £0.23 million). By comparison, the UK’s 100,000 Genomes Project received GBP £311 million.
The UK’s long term genomic healthcare implementation strategy, combined with its world leading history in the development of genomic infrastructure and data, make it better placed than many countries to explore the application of genomics beyond health. The UK may experience data and research challenges earlier than other nations and therefore gives it the opportunity to lead internationally on ethics and regulation as well as advances in data and research.
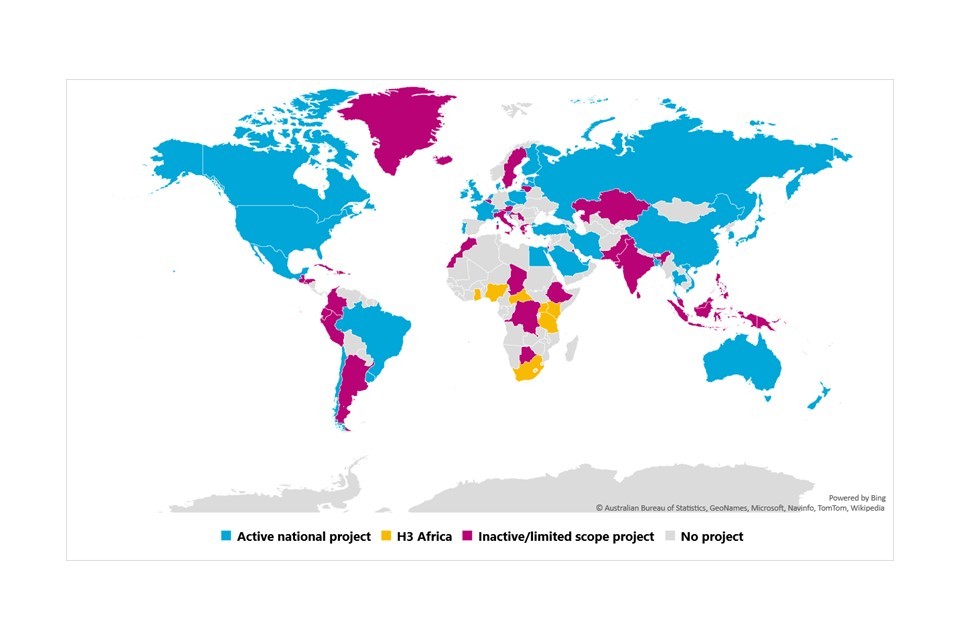
Figure 12: A map outlining the national (and some international) genomic research initiatives active in 2021.[footnote 78] Figure showing data from publication available under Creative Commons, accessible here.
Genomics and the COVID-19 response
The use of genomic sciences in epidemiology has been of increased prominence over the past year. Throughout the COVID-19 pandemic, the UK has been at the forefront of using genomic analyses to monitor the transmission of SARS-CoV-2, as well as to identify the emergence of new variants. In doing so, the UK has been able to identify more contagious and deadly variants of interest much earlier than other nations.
The impressive scale of our genomics-led response to COVID-19 can be largely attributed to the work of the COVID-19 Genomics UK Consortium. The Consortium was publicly announced on 23 March 2020, with a £20 million investment from the UK government. Membership of the Consortium is diverse, and includes the NHS, Public Health Agencies, several academic institutions across the UK, and the Wellcome Sanger Institute, which acts as a central hub to operations. Since March 2020, the combined efforts of the Consortium have led to over 800,000 SARS-CoV-2 virus genomic sequences (as of August 2021), with a large proportion of these also uploaded and shared to global public databases, ensuring that the UK is at the forefront of the genomic fight against COVID-19 (Figure 13).
The work of the Consortium is a timely demonstration of the impressive value of genomic science in public health, epidemiology, and microbiology. Indeed, the use of genomic sciences has been crucial for the development of diagnostic tests[footnote 79], vaccine development (and variant prediction)[footnote 80],[footnote 81],[footnote 82], surveillance[footnote 83], and the tracking of changes to virulence and transmissibility patterns[footnote 84].
The lessons and principles gained from the pandemic can be extended beyond COVID-19. Such principles have been the guiding force behind the recent creation of the international Pandemic Preparedness Partnership, which aims to support the expedited development of vaccines, therapeutics, and diagnostics for future public health emergencies, all of which are underpinned by genomic science. The experiences in responding to COVID-19 have also been incorporated into the recently published 2021-22 Genome UK implementation plan, which outlines the steps being undertaken to develop a ‘world leading pathogen genomics system’ for the future surveillance of infectious threats[footnote 85].
This is because genomic tools are applicable beyond viral epidemiology – they can also monitor outbreaks between humans[footnote 86], other animals[footnote 87], plants[footnote 88], or through exposure to contaminated substances such as food[footnote 89], soil[footnote 90], water[footnote 91],[footnote 92] or air[footnote 93],[footnote 94].
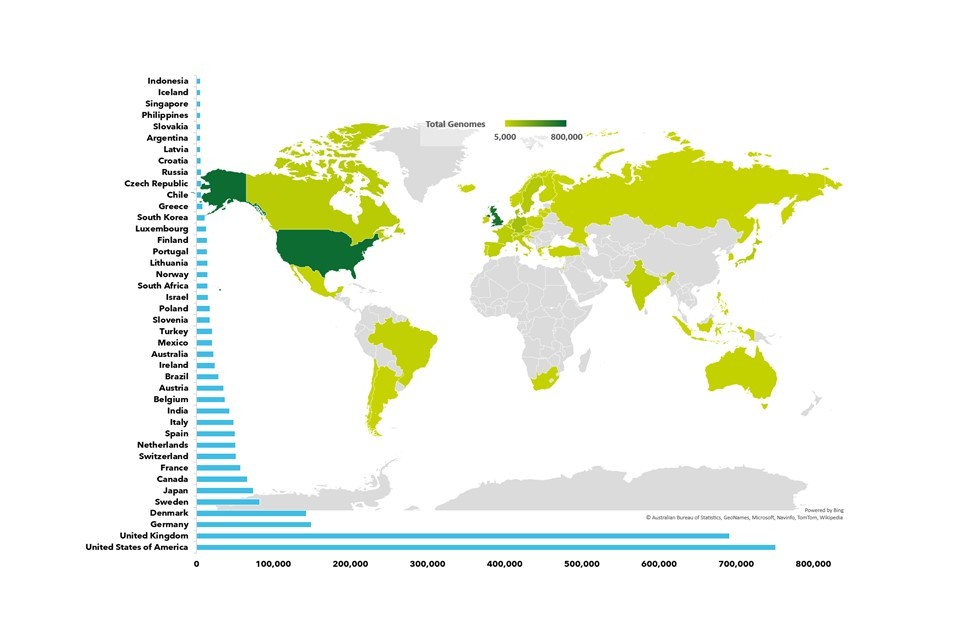
Figure 13: The UK is at the forefront of global SARS-CoV-2 genomic sequencing efforts. The data above outlines the total number of uploaded SARS-CoV-2 sequences to GISAID (Global Initiative on Sharing All Influenza Data), between 10 January 2020 and 17 August 2021. Map-plotted data excludes countries which have uploaded less than 5,000 genomic sequences.
Genomics, the microbiome, and human health
The human microbiome, that is, the collection of microorganisms which naturally reside on or in human tissues, is also important for health. Research in this area has established links between the microbiome and how a person might respond to cancer treatment[footnote 95], how the microbiome changes during pregnancy or delivery (and how this subsequently influences the baby)[footnote 96], and how the microbiome can affect or influence a person’s mood[footnote 97].
The health-focused application of microbiome-effecting strategies is currently limited to a small number of therapies given that it is still an emerging area of science. Some current examples include faecal transplantation for treating recurrent Clostridium difficile infections and probiotic dietary supplementation[footnote 98]. However, our understanding of the microbiome and the role it plays in health and disease is rapidly improving, in large part due to advances in genomic research, and these could one day translate into more novel treatments.
Recent initiatives in this area are helping researchers to investigate the roles of each individual microbial species at a genomic level and relate these back to health functions[footnote 99], and large genomic datasets. These datasets, containing the sequences of millions of microbiome-associated microorganisms, are now freely available online[footnote 100],[footnote 101]. Potential future avenues of microbiome-associated research could see clinicians using genomic-based microbiome screening to identify people with pulmonary arterial hypertension[footnote 102], treat obesity through optimization of an individual’s gut microbiome[footnote 103], or consider the interaction between anti-depressants and the microbiome to maximize treatment efficacy for psychiatric disorders[footnote 104].
Genomic medicine: summary
Understanding where genomics is making an impact today requires an understanding of its healthcare context. Genomic medicine will likely represent the first exposure of personal genomic sciences to the wider general public. The healthcare applications of genomics also demonstrate the UK’s national genomic capabilities and can therefore help to predict how applications beyond health may be developed in the coming years.
The UK’s healthcare genomics strategy, Genome UK, is highly ambitious. Few other national strategies seek to fully integrate genomic sciences within healthcare. The NHS Genomic Medicine Service (GMS) will be one of the first healthcare systems to use genomics to inform clinical decision making, spearheading the use of genomic sequencing to help those with rare diseases, cancer, and infectious disease.
The UK’s sequencing goals are also highly ambitious. By the end of March 2024, the UK aims to have sequenced one million genomes between the NHS GMS and UK Biobank (500,000 each). This will allow the UK to cement its position at the forefront of genomic sequencing data internationally.
The growth in health-related genomics has the potential to unlock genomic research and applications in other areas. The goals set out in Genome UK represent a solution to many of the current limitations in genomic healthcare research, but the pace of advancement will encourage teams across Government to seriously consider the impact of genomics research beyond health. Given the history and development of genomic data and research, the UK is in a stronger position than many countries to explore the application of genomics beyond health. As a result, the UK may experience data and research challenges earlier than other nations and have the opportunity to lead internationally on ethics and regulation as well as data and research.
Direct-to-consumer testing
Direct-to-consumer (DTC) genomic testing is generally available to the public with two advertised purposes: for ancestry or genealogical analysis, and for establishing trait or disease risk. The global market for DTC genomic testing was estimated to be USD $1.2 billion in 2020 and is predicted to grow to USD $2.7 billion by 2025[footnote 7]. Drivers behind this trend include rising awareness of genomic testing, its ease of access, customer empowerment and curiosity, opportunities for service personalisation, and falling cost of genotyping [footnote 105]. The rapid uptake of DTC testing, which is often based on DNA microarray methods, is shown in Figure 14[footnote 106].
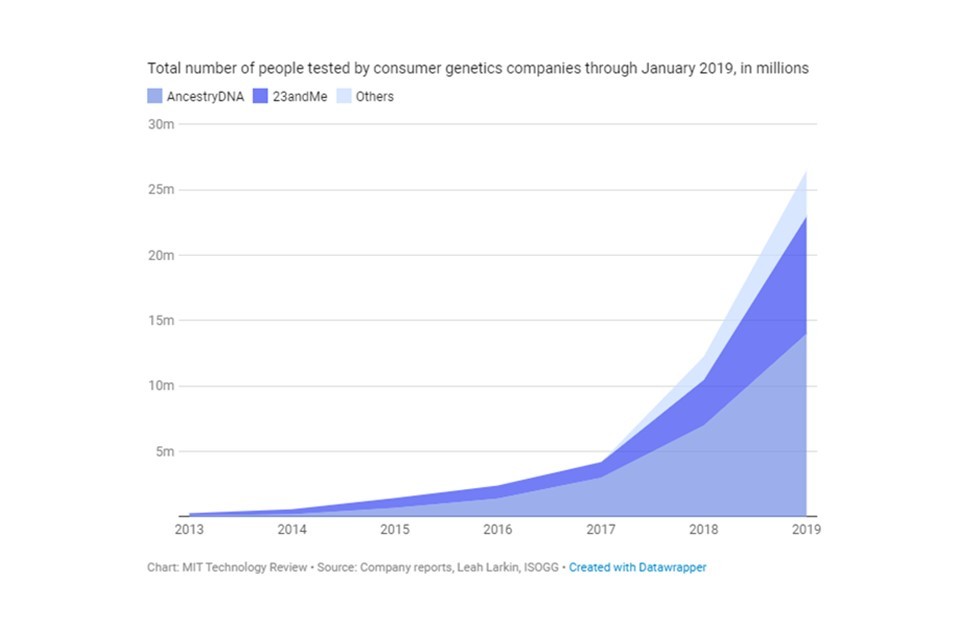
Figure 14: The growth of DTC genomic testing has rapidly increased since 2017, with AncestryDNA and 23andMe processing the bulk of DNA genotyping services. These services largely focus on DNA microarray technology to determine consumer SNPs, as opposed to whole genome sequencing.[footnote 106]
This growth raises several important questions about the information that companies provide consumers based on their genomic data. These questions relate to the strength of the data, the reliability of the analyses underpinning the provided information, the responsibilities that companies have relating to support when delivering information, data access, and the implications of holding and transferring personally identifiable, private information to international companies.
What types of DTC tests are available, and what information can they provide?
Ancestry analysis Two of the largest companies that provide ancestry analysis are AncestryDNA and 23andMe. These, like other companies, test consumer DNA for single nucleotide polymorphisms (SNP) which are strongly preserved within certain populations, broadly indicating the consumer’s ancestry.
Ancestry is a process-based concept that refers to the genealogic relationship of individuals to one another[footnote 107]. This means that the outputs of ancestry analyses generally take the form of a percentage of the customer’s genome that is associated with other individuals from broad geographical regions, such as “5 per cent Scandinavian” and “30 per cent West African”. Customers can opt-in to having this data cross referenced with other consumer users’ genealogical information for the purposes of identifying related individuals within the company’s database. This has numerous implications for consumer privacy beyond the direct consumer-company relationship, which are discussed later in Chapter 6.
Predictive testing: Some companies (such as 23andMe) may also offer predictive information for a range of traits, ranging from height to disease risk. This can encompass a mix of diagnostic or strongly predictive tests, such as for monogenic diseases like cystic fibrosis, and probabilistic tests with varying accuracy, for polygenic traits such as height. This data is obtained by comparing consumer DNA samples to known genomic indicators identified in GWAS (discussed in Chapter 3). Predictions using these tests can include determining carrier status for genetic diseases, assessing disease predisposition, or assessing predisposition for other physical characteristics (for example an ability to taste certain flavours or hereditary hearing loss). They can also include non-medical markers which may indicate physical performance capacity or exercise-related health, though the predictions for these may be extremely inaccurate (a fact which is often not clearly stated by DTC companies).
However, as discussed in Chapter 3, limitations of the GWAS which underpin these predictions mean that they are not able to detect all relevant genetic markers. Therefore, the predictions for polygenic traits only capture a small fraction of all genes relevant to a trait. For example, the estimated heritability of height is around 80 per cent[footnote 108], but polygenic scores for height can currently only explain around 25 per cent of the variation in height amongst individuals of European ancestry[footnote 3]. This may give consumers an inaccurate picture of their genetic liability or risk for a trait, especially for rare pathogenic variants[footnote 109].
Therefore, whilst DTC companies offer personalised information based on genetic research, it is not yet clear how accurate this information is. Should genomic analyses reveal potentially upsetting results, DTC companies are not obliged to offer professional genetic counselling to aid consumers’ interpretation, as is now standard in healthcare[footnote 110].
The duty of care requirement was explored by the House of Commons Science and Technology Committee in 2020 as part of their inquiry into Commercial Genomics services[footnote 111][footnote 18]. A representative for 23andMe reported that just 2 percent of users make an appointment with their GP based on their results[footnote 112].
Between December 2014 and December 2015, no complaints were raised against the company for failing to provide sufficient support to consumers to interpret their results. However, the Committee considered the issue significant enough to recommend that the government consider amending the regulation in this area. This could include requiring companies to inform consumers of the potential consequences of genomic test results for their relatives, and ensuring that companies undergo an external, independent assessment on the evidence supporting their testing offer. It could also include medical supervision or the provision of genetic counselling for some DTC genomic tests.
Several tests and services can stretch the interpretation of genomic associations beyond what the science can reliably or accurately conclude. Addressing the strength of evidence underpinning these tests, the House of Commons Science and Technology Committee issued recommendations. These were that DTC tests should undergo stricter performance evaluation prior to entering the market, and that there should be regular independent validation of the evidence base underpinning the tests[footnote 18].
Whilst the committee’s focus was on the medical applications of DTC tests, this initiative could be applied more broadly as the scope of DTC testing widens. For example, some dating companies use DNA sequencing to match consumers with potential partners, analysing a small number of genes to provide users with a purported ‘compatibility rating’. These include the human leukocyte antigen (HLA) genes, which have been evidenced to play a role in the perception of other people’s odours. Other DTC companies advertise testing designed to inform couples’ decisions about future children, by providing information about their genetic risk for certain traits or even offering pre-natal genetic screening through embryo testing. The science in this area is currently unclear and there have been calls for more responsible communication by DTC companies[footnote 113],[footnote 114]. Some companies have also used data from DTC genomic testing companies to offer personalised music playlists that are claimed to be inspired by customers ‘origins’, or ‘personalised’ travel guidance. The scientific evidence supporting the use of genomic data for these applications is questionable at best, so these products appear to be geared more towards ‘entertainment’ purposes[footnote 115].
Challenges, objections, and solutions
There are numerous impacts and challenges of direct-to-consumer genomic tests which fall beyond the objections to their scientific basis. These challenges are discussed below.
The expansion of DTC testing questions an individual’s right to privacy
A consumer DNA profile, obtained through these services, can potentially allow relatives to be identified. Consumers of some DTC testing companies can opt-in to being connected to relatives (identified through shared DNA on the company database) who have also opted into this service. Some sites allow consumers to upload a genomic profile obtained from one company in order to receive additional predictions (e.g., Genomelink[footnote 116], among other websites[footnote 117]) or for the identification of relatives (e.g., GEDmatch). GEDmatch has been used by police in the US to identify suspects and victims from crime scene DNA samples (discussed later in this chapter). The ability to upload genotypes of unknown provenance to databases by third parties has potential privacy, security and data ownership implications.
DTC testing companies may inadvertently interfere with an individual’s right to privacy by providing information regarding paternity and by testing minors. Parents are generally permitted to submit their children’s samples to DTC companies, with a 2017 study[footnote 118] revealing that none of the 43 companies contacted unambiguously excluded testing minors[footnote 119]. A report in 2010 by the Nuffield Council on Bioethics recommended that tests should only be offered to minors if they meet the criteria of the UK National Screening Committee[footnote 120], a position which was also endorsed by the House of Commons Science and Technology Committee in 2021[footnote 18].
In addition, the Human Fertilisation and Embryology Authority (HFEA) has contacted the largest DTC companies to raise concerns about the lack of signposting and support for people who might find out that they are donor-conceived[footnote 121], and have also noted the implications for donors who may have provided eggs or sperm anonymously before 2005, when donor anonymity was lifted in the UK.
Most DTC companies hold data internationally, raising data protection concerns
Most of DTC companies’ data are held outside the UK, given that they generally operate from the US. This does not in itself increase security risks, but privacy concerns have been raised about the reach of the US Patriot Act and its potential use by US security services to gain access to DNA sequence data[footnote 122],[footnote 123] including that held by researchers[footnote 124]. More recently, the 2019 CLOUD Act agreement permits US and UK law enforcement to request access to electronic data linked to serious crime directly from tech companies based in each other’s country[footnote 125], which could include those which hold genomic data of individuals under certain investigations.
The use of users’ data is generally subject to detailed terms and conditions that consumers may not in practice read closely[footnote 126], a concern that has been raised in relation to companies sharing of personal data more broadly[footnote 127]. There are also potential data security risks where companies are acquired or merged, especially when this involves companies from two or more countries. Company acquisitions or mergers may result in a change of regulations governing a company’s data use. Likewise, security risks may arise when data are transferred to other companies, either as part of the agreed terms of conditions or by customers transferring data themselves for additional testing. Data may also be leaked through lapses in cybersecurity[footnote 128],[footnote 129], raising concerns about data privacy and the potential misuse of genomic data by criminals.
The General Data Protection Regulation (GDPR) does offer some protection to consumers in Europe as it applies extraterritorially, i.e., EU rules protecting personal data continue to apply regardless of where the data is transferred[footnote 130]. Whether this is effective enough for highly personalised medical data remains to be elucidated.
The current regulatory landscape of DTC genomic testing
The global nature of scientific and commercial developments in genomics, combined with the pace of change, creates particular challenges for governance and regulation.
The scale of data gathering and portability across organisations and countries prompts concerns for security, privacy and consumer understanding of how their data will be shared. The US Federal Bureau of Investigation (FBI) is also concerned about genomic data sharing between DTC companies and other nations. In recent years, many Chinese investors have bought shares in or partnered with US-based DTC companies. The FBI see the transfer of genomic data to such nations as a risking US competitive advantage, but with longer term implications for national security[footnote 131].
Difficulty in maintaining anonymity adds to privacy concerns; an individual’s genomic data is unchanged throughout their life and can be used to infer their own identity or the identify of a close relative. Consequently, legal protection may be increasingly insufficient and require international collaboration to address[footnote 132]. This report does not attempt to summarise international genomics law or catalogue the range of relevant issues in this area, but rather aims to highlight some of the challenges most relevant to the UK.
Where used for medical purposes[footnote 133], DTC genetic tests are currently regulated as in vitro diagnostic medical devices (IVDs) according to the Medical Devices Regulations (2002[footnote 134], amended 2019[footnote 135])[footnote 136]. The regulation states that any medical test must be registered with the Medicines and Healthcare products Regulatory Agency (MHRA), who provide regulatory oversight. However, non-medical DTC genomic analyses, including analyses for paternity, ancestry, or ‘lifestyle’ factors, are not regulated according to the same framework, even though some of these latter tests may indirectly provide information on an individual’s medical status by providing raw genomic data to the consumer[footnote 137].
Distinguishing whether a genomic analysis or test constitutes an IVD is important, because this categorisation would affect whether DTC companies had a duty to provide genetic counselling alongside patient results. Furthermore, there is a danger that the results of non-IVD tests could be just as impactful as those which do fall under IVD device legislation. The results of a paternity test may be considered more substantial and impactful to the individual than a test result concluding a mild association for high blood pressure. Therefore, it could be argued that the duty of care for DTC testing companies is not as simple as the classification of the test device or process it uses.
DTC tests in the USA are covered by different pieces of legislation depending on the nature of the test. For moderate to high-risk medical purposes, DTC tests are reviewed by the Food and Drug Administration (FDA) in order to determine the validity of test claims[footnote 138]. For low risk or non-medical tests including general wellness, athletic ability or genealogy tests, DTC tests are not reviewed before they are offered to consumers[footnote 138]. In addition, many DTC genomic services originating in the USA are available in any country, and they do not necessarily obtain approval from the consumer’s country of origin. The global nature of the genomics industry makes it difficult to enforce non-US legislation to these companies in practice[footnote 139]. This means that non-medical tests based in the USA can be marketed to UK consumers with little legislative or regulatory oversight by authorities.
Non-diagnostic DTC tests aren’t explicitly regulated but do otherwise fall under several other legislation surrounding advertising, data and consumer law[footnote 140]. As a way of addressing this regulatory gap, voluntary self-regulation was proposed by the Human Genetics Commission in 2010 via the Common Framework of Principles for direct-to-consumer genetic testing services[footnote 141]. The principles were produced by a group including industry and consumer representatives. They aim to promote high standards and consistency in the provision of genetic tests amongst commercial providers at an international level, and to safeguard the interests of individuals seeking genetic testing and their relatives. The principles cover marketing and advertising, regulatory information, information for prospective consumers, counselling and support, consent, data protection, sample handling, laboratory processes, interpretation of test results, provision of results, continuing support and complaints.
However, a recent study found that none of 15 DTC companies which advertised to UK consumers complied with all the principles[footnote 142], highlighting the need for updated and legally binding principles. The House of Commons Science and Technology Committee recently called for UK government to update existing regulations for direct-to-consumer (DTC) genetic and genomic tests, and to explore the role of voluntary self-regulation as a means to facilitate data standards and sharing between commercial providers and the NHS[footnote 18].
DTC genomic testing: summary
The direct-to-consumer genotyping market is growing rapidly, with tens of millions of users across the world. Yet whilst DTC testing can provide a broad array of genomic insights into health and disease, the accuracy of genomic-based insights which underpin these services can be very low. In addition, these tests may also impact an individual’s right to privacy, both within/between families or because of lapses in cybersecurity protocols.
Contrasting genomic analyses for medical purposes, non-medical DTC genomic tests are largely unregulated as they fall beyond the scope of the MHRA and medical devices legislation. Outside of the UK, the regulatory landscape for non-medical DTC tests is equally ambiguous. Given the worldwide scope of genomic testing, there has therefore been a drive for genomics companies to volunteer themselves for more rigorous forms of self-regulation. One such self-regulation, known as the ‘Common Framework of Principles for direct-to-consumer genetic testing services’, aims to promote high standards, consistency and best practice amongst DTC genomics companies, though implementation of this framework across the industry has been poor.
Forensic science
The use of DNA analysis in forensic science has been commonplace since the invention of DNA fingerprinting by Sir Alec Jeffreys in 1984. DNA sequences are uniquely powerful in forensic science for the identification of unknown individuals because of the following characteristics of DNA:
-
In humans, every individual has a unique DNA sequence: Two unrelated humans will have roughly 6 million differences within their genetic sequence, the majority of them being SNPs. Even identical twins, who share all their DNA at conception, differ due to early development mutations[footnote 143]. Whilst conventional forensic DNA tests cannot distinguish between identical twins, the sequencing of whole genomes may make it possible to detect rare mutations which accumulate independently throughout development.
-
DNA sequence similarity is proportional to relatedness[footnote 144]: This allows human individuals to be identified via matches to even distant family members or populations. For example, mutations that are more common amongst certain ancestral populations can act as a marker to distinguish these populations from others.
-
Ubiquity and environmental persistence: most cells contain DNA (with some exceptions, e.g., red blood cells) and DNA has sometimes been shown to survive harsh conditions. For example, DNA of usable quality has been extracted from fossilised bones.
-
Database size: The amount of DNA sequence information now available for comparison and identification is growing rapidly. The more information there is, the greater the chances are of a successful match in the given context.
DNA data obtained from a sample can be used for human identification in two main ways. The first approach is by matching the DNA profile acquired from a forensic sample to a database of DNA profiles for which an identity is known. This can then be used to directly identify individuals, such as suspects and victims, or their remains, and can also be used to establish parentages, which may be useful for proving citizenship for immigration purposes[footnote 145]. The analysis and application of DNA data in this example is currently adopted by UK police forces, and the method used to perform this analysis is discussed in more detail below.
The second way a DNA sample can potentially be used for identification is by reading and interpreting the DNA sequence to reveal a person’s phenotype. This can convey information on their height, hair colour, skin colour, eye colour and facial features, as discussed in Chapter 2. However, the reliable use of genomic information for this application requires an extremely high level of understanding on how the genome relates to phenotypic features.
What DNA analysis methods are used currently for forensics?
The methods currently used by UK police are not based on whole genome sequencing. Instead, they are largely based on DNA profiles that look at areas of DNA known as short tandem repeat (STR) regions. In these STR regions, short nucleotide sequences are repeated several times, and the number of times this repeated sequence occurs within an STR is known as the copy number variation (CNV). STRs are prone to harmless errors in DNA replication, which means that they are highly polymorphic (i.e., there is a wide range of variability between people, which makes them effective for identification purposes).
By testing multiple STR sites at the same time it is possible to match DNA sequences (example shown in Figure 15). The police forensic DNA profile used in England and Wales, DNA-17, uses 16 different STR loci (giving up to 32 data points as there are two copies of each STR locus, one on each chromosome) plus a sex chromosome identifier[footnote 146]. The probability of a chance match between two full profiles from unrelated individuals is in the order of 1 in a billion (although identical twins cannot normally be distinguished). In Scotland, 23 sites are used, the DNA-17 set plus 6 others.
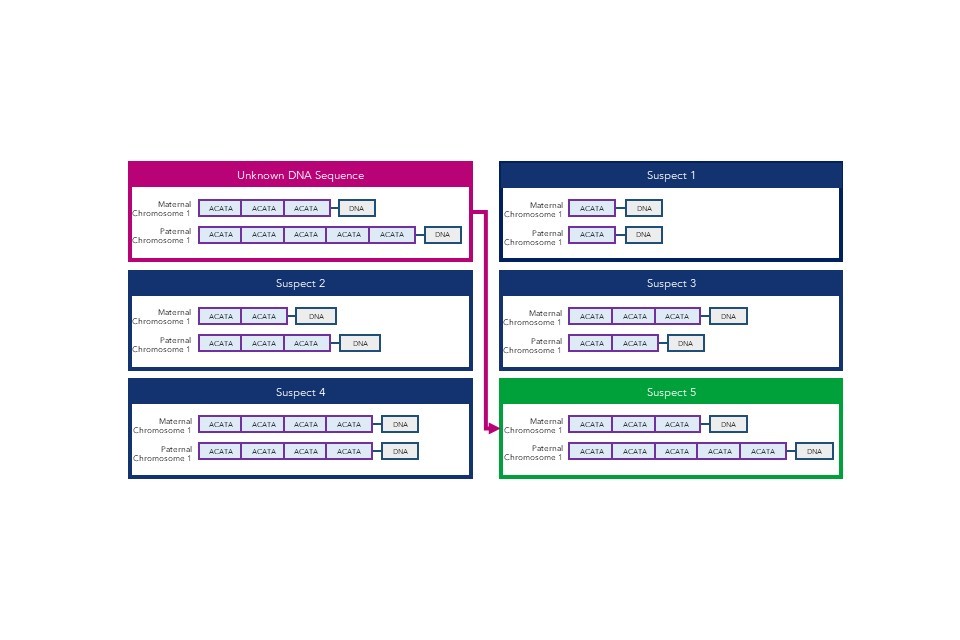
Figure 15: A hypothetical example of DNA fingerprinting. Here, analysis of a STR region on chromosome 1 identifies Suspect 5 as the DNA match based on the identical number of copy variants of the ACATA sequence.
The National DNA Database (NDNAD) holds the electronic records of DNA STR profiles (but not the full DNA sequence) of individuals and of crime scene samples from England and Wales, Scotland, Northern Ireland, Isle of Man and Channel Islands. The NDNAD was established in 1995 and since 2012, it has been run by the Home Office on behalf of UK police forces. The NDNAD provides the police with matches linking an individual to a crime scene, or a crime scene to another crime scene. However, it may only be used for detection and prevention of crime, or identification of bodies or body parts following death from natural causes – the latter was introduced following the 2004 Indian Ocean tsunami.
As of December 2020[footnote 8], NDNAD held profiles of nearly 5.7 million individuals and approximately 631,000 crime scene profile records. In 2019/2020 the NDNAD produced 23,135 matches following checks. The 2020 match rate was 66 percent (meaning 66 percent of crime scene profiles matched a subject profile already on NDNAD). Currently only STR profiles can be held on the NDNAD; full genomic sequences are not permitted. Other countries also use a core set of STRs for forensic science purposes, and the UK participates in the sharing of DNA STR data with EU and EEA countries via the Prüm Data Exchange. A mechanism also exists via Interpol that allows a member nation to search another’s DNA databases.
In England and Wales, the Criminal Justice Act 2003 allows DNA sampling on arrests for a recordable offence The power of police to retain DNA profiles has been subject to legislative changes. In 2010, the Crime and Security Act[footnote 147] established that DNA profiles and fingerprints of anyone convicted of a recordable offence would be stored permanently, while those obtained on arrest, even when no conviction follows, would be stored for 6 years, renewable on new arrests. Since the Protection of Freedoms Act 2012, those not charged or not found guilty must have their DNA data deleted within a specified period, the time depending on the nature of the offence and the age of the person.
It is also possible to perform familial searches of the NDNAD – this can identify close relatives of individual’s who were at the crime scene. By identifying relatives, the identity of the individual can be triangulated. Familial searches require approval by the Chair (or nominee) of the Forensic Information Databases (FIND) Strategy Board, and these have led to convictions in the UK in long unresolved criminal investigations (also known as cold cases[footnote 148]). 14 familial searches were approved in 2020.
Could genomic sequencing replace conventional STR methods in the future?
DNA sequencing technologies are currently not accredited for forensic use in the UK, and the NDNAD contains only STR profiles. However, genomic sequencing would provide a greater level of genomic detail regarding a forensic sample, and such analysis would provide STR information during this process.
Cellmark Forensic Services and Verogen (who now own GEDMatch) are currently undertaking validation for implementing next generation sequencing methods to provide targeted (i.e., partial) genomic sequencing capability to UK police forces. Verogen is reported to have already used genome sequencing in legal cases in the Netherlands, and some countries are currently exploring the use of whole genome sequencing[footnote 149].
If DNA sequencing were to replace current STR methodologies, the key considerations would be:
-
Cost: which has fallen considerably (though may be levelling out), and the speed of processing, which is rising.
-
Sample requirements, sensitivity, and specificity: the DNA-17 system only needs approximately 1 nanogram of DNA, whereas whole genome sequencing currently requires approximately 100 nanograms of rich template/high quality DNA (for example, obtained from a tissue sample). The capabilities of genomic sequencing approaches would also need to be evaluated to a similar standard both in general and in difficult cases, such as mixed DNA (DNA from two or more individuals) or contaminated or degraded samples. Verogen can currently perform targeted genomic sequencing on ~1 nanogram of DNA, and can sequence from 0.1 nanograms with some kits[footnote 150].
-
Operational advantages: the currently used STR equipment is highly portable, which enables rapid testing of high-quality DNA samples in the field. While some equipment for DNA sequencing is portable, ancillary equipment is still needed for analysis and careful DNA sample preparation is required.
-
Data capacity: genomic sequencing generates large volumes of data. Given that forensic data relating to serious offences has a retention period of 30 years, a challenge for adopting sequencing in this sector would be in ensuring adequate data storage capacity.
-
Whether sequencing offered new capabilities which are not available from currently employed methods at a suitable level of accuracy and precision. Examples of these capabilities include prediction of age or physical features.
-
Compatibility with NDNAD: the ability to search the existing STR database with new sequence information – as well as forwards compatibility – the ability to search a DNA sequence or SNP database with an STR profile.
-
Whether searching DTC/public databases would be permitted: this would allow deeper family searches of potentially much larger databases, increasing the chances of a match, but also raising issues of ethics and privacy.
-
The resource required for identification against a DTC/public database: investigative genetic genealogy can require substantial resource and tools to identify a familial link to sequenced DNA.
What new capabilities could genomic sequencing offer?
A significant advantage for the use of genomic technologies would be if it offered new or additional capabilities not available from current methods used within UK forensic science. The following examples are a snapshot of the potential new capabilities that genomic sequencing could offer, although all would require significant further development and thorough evaluation before implementation, including consideration of the potential ethical issues arising:
-
Prediction of physical features: Forensic DNA Phenotyping is a fast-developing subfield of forensic genetics aiming at predicting externally visible information (appearance, bio-geographic ancestry, and chronological age) from an unknown crime scene DNA sample. Pigmentation characteristics such as eye and skin colour can largely be predicted from DNA with fair accuracy[footnote 151],[footnote 152]. Height, BMI and facial structure are more polygenic[footnote 153], which makes them harder to predict with accuracy. It has been suggested that genomic sequencing could produce a “photofit” of the subject, but this technique is in its infancy and the results of recent research are contested. However, one early adoptee, Parabon Nanolabs, have early examples of such services on their website.
-
Prediction of age: DNA methylation (see epigenetics) of certain areas of the genome correlates with the age of the individual, which can be analysed with sequencing technologies, especially those of the upcoming Third Generation, which can investigate epigenetic markers simultaneously alongside standard DNA sequencing. A recent study[footnote 154] of 110 individuals aged between 11-93 years achieved less than 4 years error in prediction in 52 per cent of samples, and 86 per cent with less than 7 years.
Ethical challenges to these new capabilities
Surveillance
There are ethical challenges surrounding the potential use of DNA in criminal justice and forensics. For example, it could be possible to use DNA for the surveillance or targeting of certain groups. Recently, genomic analysis of the Uighur population was used to predict physical characteristics such as face shape[footnote 155]. Information of this type could, in theory, be used for surveillance purposes. One US company, Thermo Fisher Scientific, said it would no longer sell its sequencing equipment in the Xinjiang region of China in response to these developments[footnote 156].
Identification of individuals and privacy issues
As highlighted earlier, people can be identified through relatives’ data, even when distantly related. The recent accumulation of very large DNA databases in the DTC genotyping market has the potential to provide much more powerful searches of individuals. It has been suggested that these databases are now reaching the size at which anyone in the population can potentially be identified through triangulation of DNA data[footnote 157]. A 2019 study by Eurofins Forensic Services identified four out of ten UK volunteers using just the GEDmatch database[footnote 158]. However, this accuracy differs by ancestry, with those of European ancestry holding a 60% chance of finding a third cousin or better within the MyHeritage database, but only 40% for someone of sub-Saharan African ancestry[footnote 157]. More distant relatives that can be identified with an STR familial match, but increased uptake of whole genome sequencing could allow identification of more distant relatives than is currently possible.
Searches of DTC databases with DNA sequences obtained from a crime scene sample have been used to identify suspects in criminal cold cases. The best-known example is the Golden State Killer in the US, who was identified via multiple 3rd cousin matches to the genealogical GEDmatch database[footnote 159]. Seventy cases of murder, sexual assault, and burglary, ranging in timescales from five decades to just a few months old, have been solved across the US using genealogical approaches[footnote 160]. This technique is based on using genomic data uploaded by public volunteers to public websites. Whilst these databases do not disclose genomic data to other participants, they nevertheless facilitate genomic matching between parties. This raises legal, privacy and consent issues for commercial databases: anyone deciding to provide their DNA profile to such a database is making a choice that potentially affects their relatives, who may not have given consent. Many customers also share full family trees, including details of relatives that have not participated in DTC tests, making this identification process easier.
Law enforcement access to databases is easiest where they allow uploading of DNA by third parties. Genealogy sites GEDmatch and FamilyTreeDNA allow genotype data to be uploaded by users. By contrast, 23andMe and Ancestry.com only contain genotype data that have been generated by the company from a consumer provided saliva sample. That is, they do not permit the uploading of genotype data generated elsewhere. GEDmatch recently changed their policy for consumer consent/police access (May 2019) to require participants to opt-in to law enforcement searches, which has reduced the number of GEDmatch profiles the police may access. As of November 2019, 185,000 of the site’s 1.3 million users had opted in[footnote 160]. However, a Florida court recently granted a warrant for police to search the entire GEDmatch database including those who had not given consent[footnote 161].
Another genealogical database, FamilyTreeDNA (~2mil. profiles), permits law enforcement access “to identify the remains of a deceased individual or to identify a perpetrator of homicide, sexual assault, or abduction”[footnote 162]. It was recently reported that the company has agreed to test DNA samples on behalf of the FBI on a case-by-case basis and upload these profiles to their database. This allows law enforcement to identify familial matches to crime-scene samples[footnote 163]. 23andMe’s policy is that it will resist law enforcement access, but will comply with a valid court order, subpoena, or search warrant for genetic or personal information[footnote 164].
In response to this trend, industry guidance entitled Privacy Best Practices for Consumer Genetic Testing Services[footnote 165] was published in 2018. Developed by the Future of Privacy Forum along with leading consumer genomic testing companies such as 23andMe and AncestryDNA, it includes a ban on sharing genetic data with third parties (such as employers, insurance companies, educational institutions, and government agencies) without consent or as required by law.
Research databases
In addition to commercial databases, research databases are also of potential interest for law enforcement searches. The UK Biobank research study has stated that any attempts to use the resource for purposes other than health-related research, such as to identify participants, will be resisted. It has also explicitly stated that it will not allow access to law enforcement agencies unless forced to do so by the courts[footnote 166]. Similarly, Genomics England states that data obtained via the NHS Genomic Medicine Service is not shared with insurance companies or other government agencies, though also acknowledges that requests for data may be made by court order[footnote 167]. The Department of Health and Social Care has had confirmation from the Home Office and the Association of Chief Police Officers that they will not seek access to Genomics England’s data without a court order[footnote 168]. If a court order was obtained, then a search could be conducted by law enforcement on a UK database, as has happened in the US for GEDmatch and in Sweden for its national biobank[footnote 169]. An attempt for law enforcement access in Norway was blocked by the Supreme Court of Norway[footnote 170].
Proactive policy to prevent access by law enforcement agencies
The assurances offered by DTCs and research studies may feel insufficient to participants. To date, UK law enforcement has not sought to search direct-to-consumer or national DNA databases for forensic science purposes. Nevertheless, the circumstances under which UK law enforcement may be granted permission to search DTC or research databases merits consideration. Rather than allowing the situation to evolve by precedent through the courts, public consultation and Parliamentary debate would allow proactive policy which pre-empts access issues by law enforcement agencies.
Governance structures currently exist to regulate the use of biometric data in forensic science investigations, and these could be broadened to include genomics. While the NDNAD and DTC/research databases offer different potential forensic science uses, they were designed for very different purposes. It is important that the potential impacts on participation in health and research studies are considered when making decisions about law enforcement access to these databases.
Forensic science: summary
The current method of DNA identification within the criminal justice system does not yet use sequencing technology but relies on the detection of repeated DNA segments on certain chromosomes using capillary electrophoresis instruments. This technique does not sequence DNA but looks at specific nucleotides within a sample. This data is then cross-referenced against criminal DNA databases to find matching DNA profiles which are identical.
The increasing availability and popularity of genomic sequencing technologies has led to the development of a much richer source of genomic data which can be easily shared across databases by consumers. This data can be further annotated with family tree information and other identifying information.
This process builds a database of considerable ‘identifying potential’ outside of law enforcement, which can be used to identify relatives through triangulation of DNA. This raises potential concerns surrounding data consent, privacy and security. US and Swedish law enforcement have both already used such databases to identify individuals and solve criminal cold cases.
UK law enforcement agencies have confirmed that they will not seek access to genomic data held by Genomics England or UK research studies without a court order. However, there is currently no legislation which completely bars law enforcement from using genomic data obtained from sources other than criminal databases. Proactive policy that builds upon existing governance structures to regulate the use of biometric data to legally prohibit such access could be considered to prevent law enforcement access to these databases.
Synthetic biology
As our knowledge of genomics grows, so will our understanding of the functions of genes and how they influence different traits and systems. Improved genomic understanding will therefore enable us to make more informed decisions regarding the design and modification of an organism’s genome.
Such goals are one aspect of synthetic biology - a broad term that encompasses the creation or modification of organic material (such as entire genomes) to create new biological parts, devices, and systems, or to redesign systems that are already found in nature. This can include range from the editing of individuals genetic variants through to the creation of entire genomes. Many applications of synthetic biology are well established today, however the scope of applications is likely to rapidly develop and expand in the near future.
Current applications of synthetic biology
The applications of synthetic biology are highly diverse and have applicability across many sectors. Several applications have already been realised, with the potential for a greater range of applications in the future as genomic knowledge develops further. Examples in synthetic biology areas have included:
-
The design of novel bacteriophages - viruses that can potentially be reprogrammed to destroy antibiotic-resistant pathogenic bacteria where conventional drugs have failed[footnote 171].
-
Development of biosensors, which include synthetic bacteria that can act as a signal for detecting a target molecule. For example, bacteria which glow green if they detect heavy metal pollutants in wastewater[footnote 172],[footnote 173].
-
Engineering of non-pathogenic bacteria to treat specific diseases[footnote 174], such as an engineered strain of Listeria. This can prompt a patient’s immune system to identify cervical cancer, provoking an anti-tumour response [footnote 175].
-
Genetically modified algae, bacteria[footnote 176] and yeast[footnote 177] to produce fourth-generation biofuels. Work is also ongoing to metabolically optimise the biofuel production process, and thereby reduce carbon emissions[footnote 178].
-
Large scale manufacturing of biodegradable, microbe synthesised biomaterials[footnote 179], including cellulose, alginate and polyhydroxyalkanoates, which can be used in sustainable bioplastics.
-
Identification of naturally occurring bacteria and development of micro-organisms to aid the bioremediation[footnote 180],[footnote 181] of polluted materials such as soil and water[footnote 182]. Genomics has also helped to identify microorganisms that can naturally perform these functions.
-
Development of biocomputing: gene-edited microorganisms, such as E. coli, have already been developed into ‘living computers’ capable of executing basic computing functions[footnote 183],[footnote 184].
-
The creation of new forms of life. 2016 marked the creation of the first completely synthetic species of bacterium, known as JCVI-syn3.0. The bacterium has a small, curated genome consisting of 473 essential protein-coding genes, making it a minimal cell[footnote 185].
Methods of genomic design
The mechanisms used to generate the above examples are varied, but they fall into two main categories: editing naturally occurring genetic material or building new genetic material synthetically.
Editing naturally occurring genetic material
The process of editing a naturally occurring genome is known as genome editing. A range of techniques exist for genome editing, including CRISPR-Cas9, zinc-finger nucleases (ZFNs), meganucleases and transcription activator-like effector nucleases (TALENs)[footnote 186]. These techniques all work in a similar way; by breaking a DNA strand at a specific genetic location and allowing natural DNA repair mechanisms to seal the break. In this process, the repair mechanism can incorporate a new fragment of custom DNA or introduce a mutation to that site to render the gene modified.
Building new genetic material synthetically
The process of building DNA, chromosomes, or genomes from scratch is known as synthetic genomics. Synthetic genomics may be used to reconstruct an original, native DNA sequence, or create an entirely new genomic code. Synthetic constructs can be used to understand fundamental questions in cell biology and genomics. For example, they may be introduced into a biological host (such as E. coli) to study the effects of synthetic genomes on living systems and organisms.
Future trends in genomic design
Genome editing is faster, cheaper and relatively easier to use, meaning that its use dominates over synthetic genomic approaches. Added to this, a key limitation of the synthetic approach is the stitching of DNA segments together into longer stretches of functional DNA or a chromosome, meaning that cells from organisms which have large genomes or multiple chromosomes (such as plants or animals) are yet to be synthetically created. However, synthetic approaches may come to prominence in the near future, as synthesis costs fall and the technology to stitch genomic sequences together improves.
As of 2019, a strand of synthetic DNA costs approximately USD $0.0005 per nucleotide to build[footnote 187]. For microbes, which have smaller genomes, this was affordable enough to rapidly drive research into entirely synthetic micro-organisms – a completely synthetic E. coli genome would now cost $2300, or a Saccharomyces cerevisiae (culinary yeast) genome $6000. Coincidentally, these same two microorganisms have or are being synthetically created today: E. coli was fully synthesised in 2019[footnote 188], and work on S. cerevisiae, a multi-chromosomal yeast, is currently in progress via the Synthetic Yeast Genome Project (also known as Sc2.0)[footnote 189],[footnote 190].
Furthermore, based on these costs, building the DNA sequence corresponding to 23 human chromosomes would cost $1.6 million[footnote 191]. However, scientists estimate that by 2029 this price will fall to under $1000. Combined with advances in the tools to stitch DNA fragments together, this means that it may soon be possible to fully rebuild a synthetic human chromosome for the first time, and later, a full genome. Indeed, some scientists predict that an entirely functional, synthetic human genome may be attainable by 2045 if current rates of progress continue (see Figure 18)[footnote 192].
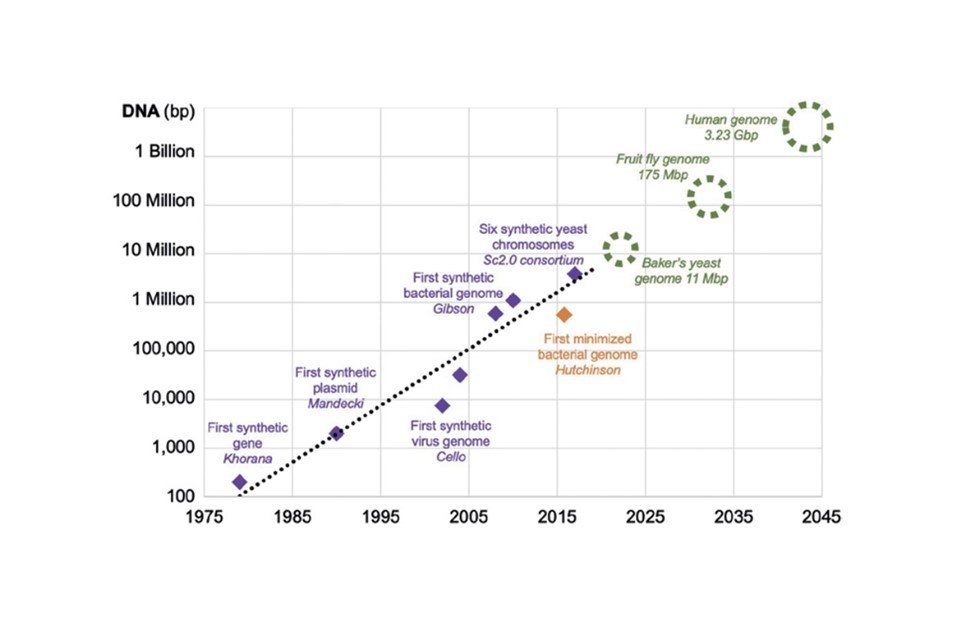
Figure 16: The progression of synthetic genomics over the past four decades. Assuming we continue the rate of progress over the coming years, we can expect to see entirely synthetic genome equivalents of yeast by 2025, fruit flies by 2035 and humans by ~2045. Figure used under CC licence from The Biochemist.
Non-biological applications of synthetic DNA
The applications of synthetic DNA can also fall beyond the biological system. In 2013, researchers at the European Bioinformatics Institute (EMBL-EBI) encoded 739 kB of data into binary format and translated this into a synthetic DNA sequence[footnote 193]. The data included a JPEG image of the laboratory, a PDF of Watson and Crick’s 1953 Nature paper establishing the structure of DNA, all of William Shakespeare’s sonnets, and a 26 second MP3 excerpt of Martin Luther King’s “I have a Dream” speech. Once the DNA was synthesized, it could be re-sequenced via standard genomic techniques, and the data extrapolated from the sequence with 100 percent accuracy. This raises the possibility of using DNA to store digital data, at high capacity, for long term preservation.
Synthetic biology and COVID-19
The scientific response to COVID-19 acutely illustrates the relationship between genomics and synthetic biology. As discussed earlier, a key to tackling COVID-19 is the widescale sequencing of the viral genome. Not only is this approach useful for identifying and monitoring emerging variants, but it also provides information on how the virus makes its proteins, and in particular, its spike proteins. Understanding the genomic sequence behind the spike protein has been crucial in the design of novel therapeutics against the virus, including for vaccinations based on mRNA. Such vaccinations work by injecting copies of synthetic mRNA, encoding the viral spike protein, into the individual. The mRNA then enters their muscle cells, and the cells translate the mRNA into the spike protein. The spike protein is then recognised by the individual’s immune system, and this confers immunity to the virus.
What impacts might synthetic biology have in the future?
Synthetic biology can feed into the future of manufacturing, as these applications may offer us a unique capability to manufacture a range of new or existing molecules and materials that would usually be too expensive or difficult to produce. Furthermore, many of these manufacturing approaches would be regarded as environmentally sustainable, as several of them essentially work by fermentation[footnote 194].
Large scale collaborative projects to build synthetic human genomes have been initiated, in recognition that it is not a case of if, but when these technologies will be developed. The largest project, Human Genome Project-Write, focuses on the design of synthetic human cells for exclusive use in fundamental biological research applications. Downstream applications of this includes disease research and work on viral resistance[footnote 195].
Synthetic biology can contribute to biodiversity. The ability to write and synthesise genomes could mean that one day it will be possible to rewrite the genomes of extinct animals, such as the woolly mammoth or the sabretooth tiger. It may also be used to restore the genomic diversity of species following a loss of biodiversity or to develop new amino acid codon sequences to generate novel proteins with novel functions not found in nature[footnote 196],[footnote 197].
The popularity of synthetic biology is resulting in a growing community of citizen scientists (sometimes known as biohackers)[footnote 198], some of which have already completed projects in the genetic engineering of simple micro-organisms[footnote 199]. Some of these groups have support from UK universities and private companies, and have received approval from the Health and Safety Executive (HSE) to operate at Class I biosafety level, in order to perform genetic experiments on microbes[footnote 200],[footnote 201].
Risks and ethical challenges
Risks
There are concerns over the risks posed by the release of synthetic organisms into the environment, whether this is deliberate or is accidental. These challenges have been faced by the GMO agricultural sector for decades, with strategies developed to mitigate the risks of accidental release. These include intrinsic measures (i.e., engineering the organism to poorly tolerate anything but a defined environment) and extrinsic measures, such as high laboratory standards and the physical containment of GMO crops.
But the diversity of synthetic organisms means an increased range of risks that extend beyond these strategies. Accidental release and poor recapture of synthetics may alter the dynamics of intra-species competition, which in turn may affect biodiversity. They may also inadvertently introduce toxins into the food chain, or their genes pass to other organisms through interbreeding. Nefarious actors could turn towards synthetic biology for harmful purposes, as is articulated through the dual use concept, for example by developing targeted bioweapons.
The availability of genome modification tools to those outside of laboratory environments also creates risk. The biohacking community culture is one of responsibility and safety which operates within the limits of local regulations and co-operates freely with law enforcement[footnote 202]. Organised groups are also run on a voluntary, informal code of ethics and responsibility[footnote 203]. Nevertheless, it is possible for lone amateur biologists to operate outside of these communities, in their own home, using materials which are now easily obtainable with little regulatory oversight and for these purposes. The risks this may pose within the context of self-administered genome editing, will be discussed later.
Ethics
Synthetic biology offers a fundamental reimagining of who can create or edit life. This raises a question from the philosophical, religious, and secular perspective, namely, do we have the right to make these changes over nature? The answer to this question may be absolute, or it may have nuance according to the scale of application, whether this be for the creation of micro-organisms, or the genomic modification of humans.
Synthetic biology also gives individuals the ability to influence the creation and development of organisms in a faster and more directive way than selective breeding and domestication. While this may represent opportunities, such as the ability to modify species at risk of climate change, there are questions around whether this should be done, particularly without yet knowing the full function of the genome.
There is potential for inequality in accessing or benefitting from this technology. Synthetic biology has a great transformative potential, particularly in the development of food sources, medicines/therapeutics, and fuels. However, they nevertheless remain expensive for most applications, and their development is driven by commercial needs and interests. This has led to some concerns that access to these resources may be reserved for the wealthy, whereas the risks will inadvertently be shared across all populations regardless of wealth[footnote 204], creating inequality.
Synthetic biology: summary
Understanding more about the genome may enable us to reliably influence the traits of an organism by modifying the genome.
-
The potential applications of this are extensive. They range from entirely synthetic life forms and biomaterials manufacture, to biocomputing, biofuels and bioremediation.
-
Some predict that by 2045 it will be possible to write and recreate a synthetic replica of the human genome from scratch, paving the way for a new level of genomic understanding.
-
The ethical implications that synthetic biology may provoke are diverse. They include concerns over inequality of access, environmental impact, the dual use concept, and the impacts that these uses may have on human health.
Agriculture and food
Whilst the future of food and farming has been explored by GO Science in a previous Foresight report published in 2011, the ways in which genomic sciences can feed into that future has undoubtedly developed since then. McKinsey & Company now estimate that the scale of the global agricultural genomics market will be equal to the healthcare genomics market by 2040[footnote 205]. This case study therefore aims to briefly explore how genomic science is helping to influence and improve agriculture in the present day and in the near future, as well as explore the public attitudes to such technologies.
Genomics and selective breeding
The global population is expected to increase to 8.5 billion by 2030 and 9.7 billion by 2050, with the majority of growth occurring in developing countries[footnote 206]. In addition, the impacts of climate change are known to affect food production, as temperature fluctuations, adverse weather events and changing water availability all decrease the yield of many key crops and affect livestock[footnote 207]. To cope with these changes, total world agricultural output needs to increase by between 60-70 per cent by 2050[footnote 208],[footnote 209]. A solution to achieving this goal includes increasing agricultural productivity – this can be through broadening environmental resilience, maximising food yields or water efficiency, or improving pest/disease resistance. Increasing productivity can be achieved in part by adapting crops and animals to suit particular applications or environments through selective breeding[footnote 210],[footnote 211].
Genomics can better inform selective breeding processes by providing more information on the species or breed of interest. Whilst crops and animals bred using traditionally selective methods are chosen based on their end performance (for example, growth rate, or meat quality), combining this data with genomic analysis, in the form of Genomic Breeding Values (GEBVs), allows producers to better understand the assets of their crops/livestock ahead of end-point production and to selectively breed them accordingly. This genomics-informed application of selective breeding is known as genomic selection.
Conceptually, GEBVs are the agricultural equivalent of polygenic scores[footnote 212]. Both approaches take genomic associations (identified through GWAS) and use these to predict phenotypes. Both methodologies have also been traditionally based on data obtained from DNA microarrays. However, there is a key difference: whilst a polygenic score is used to predict the future phenotype for an individual, contemporarily in the context of a human developing a disease, a GEBV is more of a potential breeding value. In this sense, it means that a GEBV is an estimation of what genomic influence that individual can contribute to their offspring within the context of preserving or improving the breed, rather than making a prediction isolated to the individual itself.
GEBVs allow producers to:
-
Measure traits otherwise determined at slaughter/harvest – such as meat quantity, or fruit quality.
-
Measure traits that would only be traditionally measurable in one sex – for example, male cattle can be assessed for milk production genes. This is beneficial if improvement/preservation of these genes is required within a dairy breed.
-
Provide predictive tests for livestock health and longevity – GEBVs can be performed on newly born livestock. The scores can then be used to predict the onset of genetically driven diseases before they develop, or to predict longevity.
-
Improve breed standards over time through genomic selection – for example, farmers can improve the welfare of their livestock over successive generations by breeding animals with a low genetic propensity for harmful traits, such as feather pecking or cannibalism[footnote 213]. Combined with complementary welfare strategies, i.e., optimised nutrition and housing, the combined effects would increase welfare standards over successive generations.
-
GEBVs are particularly prevalent for predicting livestock characteristics. As livestock have different genome and population dynamics, this means that GEBVs often have a greater predictive power than polygenic scores do for humans. For example, when these factors are incorporated into SNP-based heritability estimates, as much as 80-90 per cent of the genetic variance in cattle milk yield can be explained by analysing cattle SNPs[footnote 212]. Given this greater predictive power, it is estimated that rates of genetic gain (i.e., breed improvement) in common livestock can improve by as much as 30 per cent if GEBV-informed genomic selection is used in place of traditional selective breeding methods[footnote 208].
As genomic technologies become more accessible, GEBVs will become more relevant to the selective breeding process Leading the way in the adoption of agricultural genotyping is the cattle dairy industry. Between 2008-18, over 3 million dairy cattle had been genotyped worldwide, and genetic progress (i.e., increasing breed performance) in some species have doubled due to these methods[footnote 214]. Genomic selection strategies have also been shown to be beneficial to the breed standard (and often commercially available) for a range of farmed species, including sheep[footnote 215],[footnote 216], fish/shellfish[footnote 217], and a variety of crops[footnote 218],[footnote 219].
However, optimising cultivar/breed health should not come at a cost of neglecting optimal welfare standards. For example, hornless (polled) cattle could theoretically be kept in denser populations, but this would be at detriment to their available roaming space, health, and infectious disease risk.
The agricultural sector is seeing a similar shift from microarray to genome sequencing for genotyping crops/animals Sequencing approaches can boost the accuracy of trait-gene identification in organisms of agricultural interest, and thereby improve the reliability of genomic selection processes[footnote 220],[footnote 221]. This is particularly important to consider when exploring the future of genomic selection, as sequencing approaches offer greater data volume and marker identification probabilities than microarrays[footnote 222][footnote 223]. However, given the diversity of the agricultural sector, how cost-effective a GEBV-led approach to selective breeding will be for all applications is still to be determined – the most likely solution is that genomic selection will be performed on a case-by-case basis for each application depending on breeding mechanics and population dynamics[footnote 220][footnote 224].
Genomics, gene editing and agriculture
Producers can also improve breed genomes through genetic modification, and genomics is helping to drive progress in this area. Using this approach, genes of interest can be added, removed, or otherwise modified to create ‘improved’ organisms, where they are known as genetically modified organisms, or GMOs. Genomics facilitates this process by aiding the identification of genes of interest across species, and by improving the ability to predict the impacts of changes in each organism.
GMOs are one of the oldest applications of genomic science. The first genetically engineered crop was a variety of tobacco, developed as a proof-of-concept organism back in 1983[footnote 225]. Today, over 189.8 million hectares[footnote 226] are now used to grow GM crops in 24 countries, with the US, Brazil and Argentina being leading producers[footnote 227]. These crops are then exported to 70 countries for applications ranging from textile manufacture, food production and animal feed[footnote 228].
An advantage of GMOs is that genes can be introduced to the organism from other breeds or species, and without the requirement of serial breeding to maintain the original genome. This is unlike selective breeding, which risks a dilution of the breed genome when introducing a new gene variant into the breed stock. In this regard, GMOs can make the curation of breed genomes much simpler and faster when compared to selective breeding processes.
GMOs can be engineered to suit the end user and tailored to a particular environment. The most common varieties of GMO grown worldwide (as of 2021) broadly fall into one or several of the following categories[footnote 229]:
-
resistant to insect damage
-
resistant to viral infection and disease
-
tolerating of the application of selected herbicides, such as glyphosate
In 2018, nearly all soy, cotton and corn crops grown in the US were genetically modified. Together, GM varieties accounted for 94 per cent of all soy and cotton, and 92 per cent of all corn grown in the US. The US Food and Drug Administration states that most US GM corn varieties were modified to be resistant to some pests and herbicides, and cotton for pests[footnote 230]. However, future avenues for GMO engineering could focus other characteristics, such as resilience to drought or nutritional enhancement. Such developments would be facilitated by vast genomic data sets, which would help to identify advantageous genes across other species – these could then be incorporated into modern crops/livestock.
An example of this is Golden Rice, which was developed in the early 2000’s and received its first planting approval from the Philippines in July 2021[footnote 231]. Golden Rice has been modified to express beta-carotene, a precursor of vitamin A. Vitamin A deficiency affects 250 million children worldwide, is responsible for 4500 child deaths daily[footnote 232], and causes 250,000-500,000 cases of untreatable blindness each year. The consumption of Golden Rice could therefore prevent approximately 1–2 million childhood deaths per year[footnote 233]. However, moves to approve Golden Rice have been met with opposition from environmentalist groups. In their 2016 position statement, Greenpeace argued that “corporations are overhyping ‘Golden’ rice to pave the way for global approval of other more profitable genetically engineered crops” and that vitamin A deficiency can instead be addressed by promoting a “diverse healthy diet”[footnote 234].
Genetic modification differs from gene editing, although they are currently regulated in the same way Whilst plants and animal genomes can be changed through both approaches, genetic modifications arise from the transfer of genes from one species to another. For example, in the case of pest resistance, most GM plant crops contain an additional gene isolated from the Bacillus thuringiensis bacterium[footnote 235], and the beta-carotene in Golden Rice was isolated from daffodils and corn[footnote 232]. Gene editing instead uses molecular tools (such as CRISPR) to edit the existing DNA sequence of the crop or animal, without the addition of genes from other organisms.
The Department for Environment, Food and Rural Affairs (Defra) is reviewing the regulation of genetic technologies in England[footnote 236]. A key driver of their review is the classification (and thereby regulation) of gene edited organisms, as editing techniques can be used to make genomic changes to the same degree as traditional selective breeding approaches, but with a much faster speed and accuracy, and without incorporating genes from other species or cultivars as is the case with many genetically modified organisms. The divergence of legislation to recognise the differences between gene edited and genetically modified organisms could open the door to innovation in this area, and bring the UK’s legislation to be more in line with that of countries outside the EU.
However, public perception to GM foods remains polarised. Recent research by YouGov has shown that the public is equally divided on the acceptability of GMO foods (Figure 17), but that younger consumers between the ages of 18-24 have a more positive perception than older generations, particularly those aged 65 and above.
Figure 17: Public perceptions on the acceptability of GMO food. The public has a mixed perception on the use of GMOs for food production. However, attitudes vary between age demographics. Data source: YouGov.
A diversity of opinion was also recognised by Defra following their consultation on genetic technologies[footnote 236]. It found that most individuals (87 per cent) and businesses (64 per cent) felt that gene edited organisms posed a greater risk to human health/environment than conventionally bred organisms, and similar proportions (88 per cent and 64 per cent respectively) supported continuing to regulate the products of gene editing as GMOs. However, these views were balanced by academic institutions and public sector bodies, the majority of whom felt the risks were equal (63 percent, 82 percent respectively), and that they should be regulated differently (58 percent, 55 percent).
Genomics in food quality assurance
Current regulation specifies that consumers have to be appropriately informed about the food they consume, for example in respect of the ingredients used, whether they are/contain GMOs, the geographic origin and any processing the food has undergone[footnote 237]; incorrect food labelling tends to negatively affect consumer confidence, and in some cases can have safety implications[footnote 238].
Genomics can play a role in upholding food quality standards, by improving capability to monitor food composition and support enforcement of food labelling requirements. DNA-based techniques are a useful tool to identify any undeclared or mislabelled ingredients in foods, due to the accuracy, sensitivity, ease of testing and stability of DNA under a variety of food processing conditions[footnote 239],[footnote 240].
These tools have a particular power for detecting GMOs within consumer products. As gene editing techniques can produce small-scale (single nucleotide) genomic changes within an organism, there is a need for untargeted NGS-based detection methodologies which can detect them. This is because prior methodologies for detecting adulterated products were targeted approaches (such as you needed to know the genetic edit to ‘look for’ to find it within the product). This created challenges when detecting products of new or unknown mutations.
This method can also detect mislabelled produce more generally, such as non-GMO pork or beef which may be a substitute for premium priced game products; species counterfeits in canned fish such as tuna (where species differ in quality, value, and price) and substitution of cow’s milk for goat, ewe, or buffalo in cheese manufacture[footnote 241]. It has also been applied to highly processed products: a study in Brazil used NGS to investigate labelling of fish cakes, identifying mislabelling rates of 41 per cent[footnote 242]. Similarly, genomic research funded by Defra was able to differentiate between UK blue cheeses and non-UK blue cheeses based on the genomes of the fungi and bacteria present within the product[footnote 243]. Although NGS is a research tool with great potential for food quality and traceability analysis, there is a need for further research, method validation and standardisation of approaches to fully realise these applications and ensure testing methods developed are robust, reliable and fit for purpose.
Genomic sequencing can also be used to identify sources of foodborne illness. Illness can also arise from the consumption of food contaminated with pathogens from non-food sources. Such sources include animals or humans, or environmental exposure. This means that a multidisciplinary ‘One Health’ approach, which encompasses all three of these aspects within the context of health and disease[footnote 244], is effective at monitoring outbreaks. Public Health England has adopted whole genome sequencing since 2012[footnote 245], and now routinely sequences these pathogens to identify clusters of outbreaks[footnote 246], monitor outbreak progress, assess the effects of control/preventative measures, and to identify potential antibiotic resistance genes.
Agriculture and food: summary
The global population increase is resulting in increased food demand. Tackling demand means increasing food productivity, which can be achieved through improving yield, pest, or disease resistance in animals or crops.
Adapting organisms to these requirements is a fundamental aspect of agriculture, and genomics can help to refine this process further by providing more information on breed characteristics, thereby improving the selective breeding process. Known as genomic selection, this approach has been spearheaded by the dairy cattle industry and is showing growth in other agricultural applications.
Genomic selection is based on GEBVs, which can be largely thought of as analogous to polygenic scores. In this way, similar benefits of switching from microarray to full genome sequencing is also realised. As sequencing becomes more available to producers, the efficiency of genomic selection will continue to increase as the strength of GEBVs are improved. This also opens the door for the uses of GEBVs/genomic selection in other animals and crop varieties. However, cost effectiveness may limit roll out for some applications, given the variability of population dynamics and genome sizes between species.
But increasing our knowledge on species, breed or variety genomes also informs genetic modification processes. Modification of crop/animal varieties may see GMO counterparts supplant non-GMO varieties, as seen with some crop species in the US. The UK government is consulting with the public and key stakeholders regarding opportunities to improve legislation on GMO development and production, which could see greater adoption of GMOs in the near future. However, public opinion on the use of GMOs within food production remains polarised.
The breadth of views on GMO acceptability serves to illustrate the importance of food labelling and traceability to the consumer. Emerging genomic applications in this area can also be used to improve the rate at which mislabelled ingredients can be identified, as well as establish food provenance, safety, or detect the presence of GMOs in produce.
Environment and ecology
The use of genomics within environmental and ecology applications has been growing in recent years. Genomic sequencing offers new opportunities for identifying species, combating illegal trade, ecological/environmental monitoring and predicting climate change induced changes in flora and fauna.
Applications in some of these areas build upon historical genetic selection and may also use genome editing techniques. Yet in some areas, full sequence genomics do not provide necessary advantages over genetic technologies as fine-scale identification of individual organisms is not necessary. However, many research initiatives, such as the Darwin Tree of Life project, (which aims to sequence the genomes of all 66,000 known species of animals, plants, fungi and protists in Britain and Ireland) are using full genome sequencing to extensively characterise genomes, and thereby better understand how this relates to biodiversity[footnote 31].
Identification of invasive or protected species
Invasive species are an increasing problem in the UK and elsewhere[footnote 247]. Globalisation facilitates their arrival, while environmental changes such as climate change facilitate their establishment. Invasion can cause long-term damage for the environment (e.g., reduced ecosystem stability and loss of native species)[footnote 248] and has been estimated to cost the global economy billions of dollars each year[footnote 249].
Species status is commonly identified using a process called DNA barcoding. DNA barcoding compares sections of DNA from a sample to a reference library, such as the Barcode of Life Data[footnote 250]. Because identification is focused on the differences between species rather than the differences between individuals, DNA barcoding only need use sections of DNA rather than the full genomic sequence.
Combatting illegal trade
Genomics has also been used to combat the illegal trade of flora and fauna. The largest ivory seizure since the trade ban of 1989 was geographically traced to an origin in Southern Africa using DNA extracted from elephant tusks[footnote 251]. This enabled law enforcement agencies to restrict their investigation to a tighter area and collection of trade routes.
The trade of illegally sourced wood contributes to unsustainable deforestation which subsequently threatens global biodiversity[footnote 252]. Trade of the most threatened tree species is restricted by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), an international agreement signed by 183 countries. DNA barcoding is restricted to identifying species and is unable to accurately determine the source of species or enable accurate global tracking[footnote 253].
Phylogeographic methods look at genetic variation within a species, allowing the source region of flora to be determined. Population genetic assignment further allows the identification of individual flora from different populations in the same region[footnote 253].
Both phylogeographic and population genetic assignment methods are suited to determining the source of flora but cannot be used for tracking. DNA fingerprinting can be used to distinguish individual flora within a population within the same region, and as such it offers the ability to track flora along a supply chain[footnote 253].
For example, DNA fingerprinting has been used to identify whether timber has been substituted with illegally harvested logs at certain stages of the supply chain[footnote 254]. It was also used to identify who had illegally harvested big leaf maple in Gifford Pinchot National Forest in Washington, US in 2015[footnote 255]. Planks seized at a sawmill in the state were matched to the tree stumps from which the timber had come, leading to prosecution[footnote 256]. The Forestry Stewardship Council, US Forest Services and Kew Gardens have established the Global Timber Referencing Project to provide a database of georeferenced samples that can be used for identification[footnote 257].
Detecting contamination of water
In recent decades, increases in human population growth and urbanisation have put pressure on freshwater resources for human recreation and consumption. Contamination by sewage, toxic chemicals, nutrients, and resultant harmful algal blooms can make water unfit for human consumption or recreational activities.
Sequencing-based approaches are increasingly used for water quality monitoring alongside traditional methods such as the culturing of bacteria[footnote 258]. Sequencing-based approaches can detect the presence of faecal indicator bacteria, such as Escherichia coli (E. coli), and the presence of bacteria containing antibiotic resistance genes. They can also detect species enriched by the presence of harmful chemical pollutants and pathogens such as norovirus. Sequencing technologies are also being applied to the systematic understanding of the ecology and control of harmful algal blooms. Development of portable sequencing platforms offer the prospect of near-real time sequencing in the field.
Ecological monitoring and conservation
DNA sequencing can be used to identify protected species and groups of species of conservation interest by using DNA samples collected from animal’s environmental surroundings. This may come from samples such as faeces, hair, shed skin or carcasses[footnote 259]. These methods can also be used to estimate population sizes, offering a non-invasive approach to ecological monitoring[footnote 260]. Because there is no direct sampling of animals, this approach can be conducted on a cheaper and larger scale than traditional sampling.
For example, Natural England used DNA sequencing of environmental samples to detect the presence of great crested newts across 7,518 ponds in England[footnote 261]. These data are openly available, allowing local councils to take a more strategic licensing approach to species conservation and support local planners in making informed decisions on development[footnote 262].
Species modification
Genetic modification tools such as CRISPR could be used to control populations or to make them extinct. This has been an area of recent debate relating to species that are invasive or act as disease vectors[footnote 263],[footnote 264],[footnote 265],[footnote 266]. For example, ‘gene drive’ approaches can be used to insert genetic modifications which are more likely to be inherited as species reproduce. One study demonstrated that gene drive mutations to a fertility gene in mosquitoes entirely crashed the population[footnote 267].
While gene drive changes are currently only suitable to certain species and can be reversed[footnote 268], these technologies are in their infancy and present ethical and moral challenges relating to biodiversity.
Ecosystems and biodiversity
The Earth is changing in ways that pose significant threats to ecology and the environment. Anthropometric global climate change is driving environmental changes at a rate not seen in the Earth’s history. For example, Figure 18 illustrates the rising levels of carbon dioxide over millennia, which has been attributed to human actions. This is changing environments in such a way that they are out of the habitability limits of resident species[footnote 269]. Species may respond by either migrating or evolutionarily adapting to the changing environment.
Figure 18: Atmospheric carbon dioxide level in parts per million (PPM). Plotted data from the US National Oceanic and Atmospheric Administration.
Genomic approaches may offer potential for modelling species’ evolutionary environmental adaption potential and predict their responses to climate change induced habitat change[footnote 270], [footnote 271]. Knowledge of the genomic basis of evolutionary adaptation may help to better understand how species can adapt[footnote 272].
For example, studying the variation amongst populations across climatic gradients could provide insight into how much genetic change may be expected under different climate change scenarios[footnote 270]. This may be particularly for ‘keystone’ ecosystem species; those species most predominant in an ecosystem or that have a disproportionately large effect on its environment.
Genomic analyses into the genetic variation within species and their population size may also be used to estimate their vulnerability, future migration, and potential adaptation to climate change[footnote 273],[footnote 274],[footnote 275]. Knowledge of genetic variation that is associated with abiotic factors such as topography and environmental/climatic gradients may offer potential intervention options. One such intervention would be selecting flora which carry genotypes beneficial to climatic conditions anticipated in the future[footnote 276]. Genomic analyses may also offer opportunities to monitor plant and animal health and detect potential threats of new or invasive pathogens. This has been demonstrated with the ash dieback pathogen, where researchers have identified genes related to resistance against the disease[footnote 277][footnote 278].
Genomics may also offer promise for restoring ecosystems and biodiversity through gene editing to generate novel genotypes. Genetic modification has been previously used to create crops that are more resistant to drought[footnote 279]. However, altering species by gene selection or gene-editing poses risk to biodiversity and conservation[footnote 280].
Regulation
Whilst existing legislation is in place[footnote 281], it is important that regulatory frameworks are updated and maintained to effectively oversee and license the use of gene editing, and to monitor the introduction of gene selected organisms around the world. The risks of introducing such organisms into ecosystems are not comprehensively understood, meaning that there could be potentially deleterious knock-on effects on the environment and ecosystems and global or local conservation efforts[footnote 282]. A risk assessment framework which uses the precautionary principle to address inadvertent potential ecological, genetic or health effects would allow for effective regulation of the use of genetic applications in the environment and ecology.
Environment and ecology: summary
There are a range of genetic applications currently used within environmental and ecology fields to identify and track species, monitor the environment, help identify species that may be vulnerable to global climate change, and improve conservation efforts.
Advances in genomics has the potential to improve upon current genetic applications in environmental and ecology fields. Developments in portable technologies may improve monitoring in hard to reach and low-income areas. Developments in genomic sequencing may also be useful for better predicting species’ responses to global climate change and monitor environmental damage.
Development of international regulation around the use and introduction of genomic editing is required to manage the potential risks surrounding environmental and ecological applications.
The future of genomics: potential applications
The future of genomics is exciting – as technological advances speed up and become cheaper, there will be a huge variety of potential ways that genomic information could be applied in different sectors. Some of these are already viable, but the majority are currently ideas, or in the early stages of development/experimentation. It is worth considering both the viable and the theoretical from a hypothetical perspective, even where they may seem either unrealistic and/or undesirable, as part of good horizon-scanning practice, and to consider the need for future-proofing.
This chapter discusses some potential predictive applications of genomic technologies in five human-focussed case study areas: employment; sport; education; criminal justice and insurance. In each case, we cover what genomics is currently able to tell us about relevant traits and why this may be relevant for future applications of the technology within a predictive capacity. Some degree of reasonable uncertainty is inherent in this assessment as the science is developing rapidly. Each section therefore covers traits which may become tractable, the state of the science on these traits, and their potential implications. The chapter then concludes with a hypothetical overview on the use of genome editing, how this might interface within these non-health contexts, and what these applications might look like.
It is crucial to emphasise that the lack of data diversity in genomic databases will pose a challenge to the equitable use of predictive genomics in these fields. The DTC genomic testing market is governed by voluntary codes, operates across national borders, and there is no universal definition of when a test is sufficiently predictive. Given this, some of the potential uses of genomics considered here run ahead of what is justified by the science and are necessarily more speculative.
Key messages:
-
Advances in genomics offer promise for applications across a range of fields beyond health, such as in sport and workplace safety.
-
Policymakers and practitioners will need to react to emerging developments in genomic science in two modes. First, they will need to address the technical and ethical issues of genomic prediction to make the best use of new knowledge. Second, given the DTC genomic testing market is likely to outpace the science, policymakers will need to consider what mitigations and support should be in place to help both customers and service providers to navigate this new marketplace.
-
In many non-health fields, the science is less advanced and the operational implications more challenging than in some areas of health. For example, genomic prediction of monogenic disease is far more accurate and prescriptive than genomic prediction of complex non-health traits such as educational attainment.
-
The limitations of the current evidence base should be acknowledged. The underrepresentation of genomes from non-European ancestry individuals in genomic databases impacts on the applicability and accuracy of predictive applications. This must be addressed before applications are deployed, or they will risk entrenching inequalities in key areas of life, such as employment and education.
-
Regulation of genomic technologies in non-health fields is currently patchy, and risks being outpaced by technological advances. Proactive regulation may help to ensure that genomic technologies are not misused in non-health fields.
-
Rapid advances in genomic technology mean that consumer protection laws do not adequately govern genomic testing companies. Genomic prediction is currently in its infancy, yet the self-regulation of DTC genomic testing companies means that the limitations of their predictions are not always clearly explained to the customer.
Employment
-
Health and safety
-
Predicting soft skills
-
Genetic discrimination
Sport
-
Athlete pre-selection
-
Health screening
-
Gene doping
Education
-
Academic prediction
-
Targeted interventions
-
Learning environment
Criminal justice
-
Disinhibition and substance abuse
-
Genomic evidence at trial
-
Fairness and inclusion
Insurance
-
Voluntary self-regulation
-
Information asymmetry
-
Public resistance
Genome modification beyond health
-
Genome editing in disease
-
Modificaiton of non-health traits
-
Issues, ethics and risks
Employment
The rapid growth of genomic sciences could prompt employers to use genomics in a number of ways, such as the selection of workers of optimal health or personality for a role to prevent workplace injury. This concept has been recognised for quite some time. In 1938, British-Indian scientist John B. S. Haldane observed the health of manual labourers in Staffordshire, and noted that[footnote 283]:
“The majority of potters do not die of bronchitis. It is quite possible that if we really understood the causation of this disease, we should find out that only a fraction of potters are of a constitution which renders them liable to it. If so, we could eliminate potters’ bronchitis by regulating entrants into the potters’ industry who are congenitally exposed to it.” This chapter will first discuss how genetics and genomics can be used within employment scenarios today. It will then explore the genetic basis of many traits and characteristics relevant to employment and how these could be used to underpin applications of genomics in this area in the future.
Key messages and considerations for policy
-
Genetic testing today is only used in very limited circumstances in employment, such as occasionally in professional sport. However, many traits relevant to employment have a genetic component. This includes intelligence, extraversion and susceptibility to certain injuries.
-
As our understanding increases, the potential uses for genomics in employment will grow. This could include better understanding employee risk profiles at an individual level, allowing for mitigation measures. However, polygenic scores are not yet, and may never be, of sufficient quality and reliability to make predictions at an individual level.
-
More contentious would be the use of genomics in assessing potential for candidate selection purposes. This would raise serious ethical issues that would need to be worked through, including the potential for reinforcing structural inequalities which then are then reflected in polygenic scores.
-
There are major technical barriers to either of these uses, including poor predictive power and easier (non-genetic) testing. However, attempts to use genomics could run ahead of the science.
-
Although there are no examples in the UK to date, the use of genomics is not completely far-fetched. Indeed, there are examples in the US of companies trying to use genomics to avoid paying out compensation for injury.
-
Unlike in the US, there is nothing explicit in UK legislation barring the use of genetic testing in job or candidate selection.
Genomics in employment today
Genetic testing is already used within a very small number of workplaces For example, genetic testing for heritable heart conditions, whereby physical exertion can cause sudden death, takes place in some sports industries (see the sports case study for further information). However, outside of this narrow example, genetic or genomic testing in employment is not widespread, even where there could be potential benefits to employees in terms of avoidance of injury or death.
But testing beyond this example is prohibited or highly restricted by employment regulations and the use of the results in employment decisions could constitute unlawful discrimination. However, genetic testing through direct-to-consumer companies is now being offered by some employers[footnote 284] as an employee benefit. This focuses on the opportunity to assess personal risk for diseases such as cancer or hereditary high cholesterol, rather than job-related characteristics or health insurance purposes. The aim of these employee benefit approaches is to encourage preventative action in health, which may have benefits to the individual in terms of overall healthcare costs and outcomes. This may in turn benefit employers, even though they do not directly receive the employees’ test results to make judgements relating to suitability for employment.
Looking to the future: what else could genomics tell us?
In the future, genomic science might be able to reveal information about an individual’s employment prospects and suitability. This is because many behavioural characteristics that could be deemed relevant to work and employment are influenced by a person’s genome[footnote 11]. These characteristics include (but are not limited to) personality traits, such as extraversion (53 percent heritable), neuroticism (41 percent), agreeableness (41 percent), conscientiousness (44 percent), openness to experience (61 percent) and intelligence (considered further in the education case study).
GWAS have been conducted on a range of these characteristics, identifying some of the specific genetic markers that underlie them[footnote 285]. Currently, genetic research into academic attainment and cognitive ability has been outpacing that for other cognitive and behavioural characteristics such as personality traits.
In addition to cognitive and behavioural characteristics, physical characteristics can also play a role in work and employment. There have already been genetic studies into physical characteristics relevant to employment, including:
-
Susceptibility to injury, relevant to manual occupations. Research has found genetic influences for injury susceptibility which explain around a quarter of the variance in injury involvement for 33–60-year-olds[footnote 286]. The genetic influence is higher in older age groups, which may reflect recent changes in environmental factors that influence injury throughout the life course.
-
Bone mineral density, relevant to active occupations, such as the armed forces. One GWAS has identified nearly 900 chromosomal regions[footnote 287] associated with bone mineral density. When these markers were incorporated into a genetic algorithm, individuals who scored poorly had a 1.9 times increased risk of stress fracture.
-
Physiological functions, relevant for shift work. GWAS have identified a genetic variant related to melatonin signalling, which plays a role in circadian rhythms and sleep patterns[footnote 288]. This may contribute towards adaptation to shift work and job-related exhaustion.
-
Chemical sensitivity, relevant to some hazardous occupations. Research has identified genetic differences between people who are sensitive to certain chemicals[footnote 289],[footnote 290]. Studies have also identified genotypes which may make individuals more susceptible to cancer if exposed to heavy metals, cooking fumes and cigarette smoke[footnote 291], or asbestos[footnote 292].
Some of these characteristics are highly polygenic and therefore the partial genetic risk for the characteristic would be predicted using polygenic scores. As the reliability of polygenic scores improves, it might become possible that scores for these traits could be used in the context of employment and recruitment.
However, a few technical limitations exist that caution against the use of genomic testing for some traits. To recap, these limitations are:
-
Poor predictive power: polygenic scores are an approximate measure of an individual’s genetic predisposition towards a trait and do not provide accurate predictions for any complex traits. Furthermore, polygenic scores capture only a fraction of an individual’s total genetic predisposition to a trait. Direct-to-consumer genomic analysis, which usually focuses on common gene variants, may be especially limited in this context. These limitations restrict the predictive power of many genomic predictions.
-
The availability of effective non-genetic alternatives: for behavioural and medical traits, genetic tests may have lower predictive power than conventional tests or using family history. Examples of these conventional, non-genetic tests could include an IQ test with respect to cognitive ability, an electrocardiogram (ECG) with respect to a heart condition, or a personality test.
Therefore, the development and use of genomic testing in employment is currently very limited by current scientific knowledge. Nevertheless, there exist a number of regulatory considerations for genomics in employment that proactive policy could look to pre-empt.
Regulation
Given the challenging ethical and technical considerations involved in this area, legislation and codes of practice generally do not support genetic testing in the context of employment.
While the EU Charter of Fundamental Rights (Article 21.1) prohibits discrimination based on ‘genetic features’ under EU law, in the UK there is no specific prohibition of genetic discrimination in domestic law. The Equality Act 2010 restricts what employers can ask in pre-employment checks. Whilst genetic discrimination is not explicitly covered by the Act, some traits may overlap with other protected characteristics, which are covered[footnote 293][footnote 294].
The UK Information Commissioner’s Office (ICO) Employment Practices Code[footnote 295], published in 2011 (currently under review) also addresses the use of genetic information in employment. Its key points are:
-
Employers should not use genetic testing to obtain information that is predictive of a worker’s future general health. Obtaining employee information through genetic testing is considered too intrusive, while the predictive strength of such tests are currently insufficient, making them unreliable for this application.
-
Employers should not insist that a worker discloses the results of a previous genetic test. It is important that workers are not discouraged from taking genetic tests that may be beneficial for their health. Employers can ask for information that is relevant to employee health and safety, or other legal duties, but the provision of the information should be on a voluntary basis.
The Code recommends employers seek genetic testing only where a worker with a detectable genetic condition is likely to pose a serious safety risk to others, or where a specific working environment or practice might pose specific risks to workers with particular genetic variations. In these circumstances, the code offered the following recommendations to employers:
-
To only seek information through genetic testing as a last resort. Employers may use genetic testing where it is the only reasonable method to obtain the required information. An example of this is where it is not practical to make changes to the working environment or practices to reduce a risk to all workers.
-
Employers should inform the Human Genetics Commission (HGC) of any proposals to use genetic testing for employment purposes. (Note – the HGC was abolished in 2010, publishing its last paper in 2012. It is unclear whether this guidance will be updated in the upcoming refresh of the Employment Code.)
Furthermore, if a genetic test is used to obtain information for employment purposes, employers must ensure that it is valid as a test and is subject to assured levels of accuracy and reliability. Employers should also ensure the results of any genetic test undertaken is always communicated to the person tested, and that professional advice is available to confirm correct interpretation of test results.
Given the abolishment of the HGC body that governed this code over 10 years ago, there is a case for government to review whether this should be included in Employment Codes and assess whether oversight by another body is required.
Genomic science has progressed greatly in the past decade, making outdated regulation a potential area of risk for UK employees.
Ethics
Genomic testing could be used by some employers to prevent injury or harm to employees, and this is likely to be the least problematic use of genomics in employment with regards to ethics. As genomic science develops, with potentially more behavioural or cognitive characteristics able to be tested or predicted from genomic data, new and challenging ethical dilemmas will arise.
Using genomics to predict skills and characteristics could be discriminatory against certain employees, particularly if tests are used in isolation without additional metrics such as interviews or personality tests. The limitations of genomic research could introduce bias to employment processes using genomic data. This would most notably be due to their sampling being heavily reliant on subjects of European ancestry and the incomplete genomic data provided by microarray data as discussed in Chapter 3. However, the quality of genomic data is improving rapidly. Larger studies of individuals from diverse ancestries and the adoption of sequencing technologies will likely help to improve the applicability of genomic analysis for employment.
In the US, some employers have used genetic testing to avoid paying disability benefits or financial compensation arising from work-related injuries or illness where the tests have shown that employees had a proclivity for a related condition.[footnote 296] The Genetic Information Nondiscrimination Act (GINA), enacted in the US in 2008, has since prohibited the use of genomic information in job hiring, redundancy, placement, or promotion decisions.
As of 2021, there have been no cases of UK employers using genomic data to influence employment decisions. However, it is not difficult to imagine in future, and there is no explicit legislation barring employers from doing so.
Sport
There is significant research interest into the role that genomics might play with regard to physical and psychological traits related to sporting ability. Genomic testing has the potential to identify athletic predisposition in two ways. First, it may be used for athletic selection by identifying genetic traits associated with sporting prowess. Second, it may be used to identify individuals who are at risk of injury or death from certain activities. At present, there is no evidence that genomic information is used in the UK in this way.
Nevertheless, testing for these factors could theoretically form the basis of complex selection criteria for elite development programmes, though at present this is not supported by scientific evidence. Such an approach could result in a greater focus on early identification of perceived genetic sporting advantage. Tests could also potentially be used to tailor training or nutrition programmes to improve performance or reduce injury risk. Individuals would also undoubtedly like to know more about their genetic potential for sports performance. Genomic tests for sports science are currently being marketed to consumers, despite slim evidence supporting these applications.
Key messages and considerations for policy
-
Genetic tests can be used to screen for some health conditions that would pose a risk to athletes, and for sex verification purposes. How this informs subsequent decisions about participation in sport remains controversial.
-
Direct-to-consumer genomic tests to identify athletic potential or inform training regimes are not considered as being accurate or useful at present, and their use is discouraged by prominent sporting bodies.
-
Alleles of certain genes (including ACTN3, ACE, GALNTL6 and EPOR) have been associated with elite athletic performance. In the future, gene editing techniques could potentially enhance the performance of people whose genome does not include the advantageous alleles.
-
Regulation of genomic science in sport is limited, though the World Anti-Doping Agency has pre-emptively outlawed gene doping and is developing direct and indirect approaches to detect for its misuse within sport.
-
Genomics in sports raises similar ethical dilemmas to other applications, including those around privacy, consent, opportunity, and discrimination.
Genomics in sport today
Single gene genetic tests can be used to exclude participants at risk of harm, though alternative screening methods are more efficient. These tests are primarily for rare monogenic conditions where exertion could result in sudden death, such as hypertrophic cardiomyopathy.[footnote 297] However, these conditions can be detected by more conventional methods, such as Magnetic Resonance Imaging (MRI) scans, making a genetic test unnecessary. Italy was the first nation to use electrocardiogram (ECG) exams to screen athletes in 1982, and it is estimated that under this strategy nearly 90 percent of potential sudden cardiac deaths are prevented.[footnote 298]
Genetic testing can be used for sex verification purposes, though the wider issue of sex verification in sport is highly controversial. High levels of testosterone are regarded as providing competitive advantages in sex-segregated sport, particularly in female or intersex athletes. The rules of World Athletics state that competitors with a Difference of Sexual Development (DSD) must seek verification of their sex[footnote 299], which can be done through genetic testing. The controversy surrounding Caster Semenya’s performance at the Berlin World Championships in 2009 raised issues around genetic testing for sex verification, including the need for counselling, full information, consent, autonomy, and confidentiality[footnote 300]. Sex verification itself is regarded as conflating issues of sex, gender, anatomy, and identity, regardless of the method used[footnote 301].
There has been interest in the possibility of using genomic screening for identifying athletic talent and potential. In 2008 it was reported that at least one UK football club had explored the possibility of screening players’ genomes. This was considered to identify sporting potential through the presence of genetic variants associated with increased athletic performance[footnote 302]. A study found that UK athletes and support staff would generally be interested to know whether athletes possessed genetic variations associated with performance (81 percent and 61 percent respectively) and risk of injury (85 percent and 78 percent)[footnote 303]. The study also found that athletes and support staff would not use genetic information to determine selection or employment (28 percent and 14 percent approval respectively). However, opinion was mixed on whether it would still be valuable information for talent identification purposes (67 percent and 48 percent approval from athletes and support staff respectively).
Whilst there are no known examples in Europe or the UK, genomic screening programmes for identifying sporting potential have been reported in China[footnote 304] and Uzbekistan[footnote 305]. In 2018, the South China Morning Post reported that the Chinese government and the Chinese Academy of Sciences had planned to use whole genome analysis to inform the athlete selection process for the upcoming 2022 Winter Olympic Games. Similarly, in 2014, scientists at the Academy of Sciences of the Republic of Uzbekistan announced they would begin genomic screening of Uzbek schoolchildren to identify genetically-driven athletic potential.
The increasing popularity of direct-to-consumer (DTC) tests also raises key issues for predicting athletic potential based on genomic analysis. In 2015, the International Federation of Sports Medicine (FIMS) strongly criticised DTC companies that offered genomic predictions of athletic potential targeting children and their parents, stating that “genetic tests have no role to play in talent identification or the individualised prescription of training to maximise performance”[footnote 306]. An update in 2019 highlighted the risk that misplaced confidence in DTC genomic tests for sports could “lead to incorrect decisions such as inappropriate early specialisation for sports, inappropriate training, genetic discrimination and even increased health risks”[footnote 14].
The Australian Institute of Sport also stated in 2017 there were “currently no scientific grounds for the use of genetic testing for athletic performance improvement, sport selection or talent identification”.[footnote 307] Their primary concerns were the lack of validation and replicability of the results, along with the lack of medical counselling during the process. Whole genome sequencing may improve the predictive power of genomic tests in the future, though whether this improvement would be enough to prompt a re-evaluation of the role of genomic testing within sport is uncertain.
Looking to the future: what else could genomics tell us?
The potential applications of genomic science within sport go beyond the early selection and prediction of athletic performance. Since 1997, at least 155 genetic variants have been linked to elite athlete status[footnote 308][footnote 309]. These markers include variants associated with skeletal muscle mass, growth and function, dynamic response to training, cardiovascular efficiency, and energy metabolism[footnote 14]. This may provide a foundational basis for future athletic potential genetic screening procedures.
As knowledge of the genome increases, so does the development of gene editing techniques. One of these techniques - CRISPR - allows DNA to be cut at specific locations of the genome allowing for changes to be made through the introduction of mutations or replacement of the cut genetic material. While the science of genome editing is currently in its infancy, these techniques could make it possible in the future for athletes to engage with gene doping. This could hypothetically enhance athletic performance by editing genomes to hold advantageous alleles of key performance-related genes.
Two of the best-studied genes amenable to CRISPR and which are associated with athletic performance are ACTN3 and ACE.[footnote 310] The ACTN3 gene encodes for the protein actinin, which is involved in generating contractile force within muscle fibres and has been associated with power output. However, the R577X variant in the ACTN3 gene, sometimes called the “speed gene”, accounts for only 2 percent of variation in muscle strength or sprinting.[footnote 311] ACTN3 may also have a wider role in enhanced improvements in strength, protection from eccentric training-induced muscle damage, and sports injury.[footnote 312] However, many people already have this variant and would not benefit from gene editing.
Certain alleles of the ACE gene are associated with endurance, being more prevalent in elite endurance athletes[footnote 313] and high-altitude Nepalese Sherpas.[footnote 314] The alleles many also assist in heart rate recovery after exercise[footnote 315], and so could be a desirable genetic edit.
There are some very rare single-gene variants that may give an unusually strong predisposition to elite performance. One example is a variant of the EPOR gene, which is associated with an elevated red blood cell count (erythrocytosis). This variant leads to the production of around 25 to 50 percent more red blood cells than usual.[footnote 13] The Finnish cross-country skier Eero Mäntyranta had the EPOR mutation which probably contributed to his total of seven Olympic cross-country ski medals.[footnote 316]
Only a limited number of GWAS have been carried out with respect to elite sporting performance. The number of elite athletes is small, limiting the statistical power of large-scale genomic studies. Studies based on dichotomous groups (for example, elite athletes versus others) generally have at most a few hundred elite athletes to compare to the general public. This is far short of the number needed to reliably identify genomic variation associated with performance characteristics.[footnote 309]
A recent GWAS examining elite endurance runners illustrates this difficulty. 375 elite runners were compared against control individuals at 195,000 genetic markers, but the study failed to identify any markers that were associated with athletic ability.[footnote 317] A further study sought to examine 45 candidate markers in other groups of athletes. One variant of the GALNTL6 gene was found to associate with anaerobic performance and strength athletes, providing 5 to 7 percent higher absolute and relative mean power than those without the variant, though by what mechanism remains unclear.[footnote 318]
Further studies in this area have tried to mitigate these limitations by narrowing the scope of study and performing GWAS on specific biological factors that may contribute to overall athletic success. However, these studies are less informative for predicting sporting ability as specific factors only provide a small influence on overall success.[footnote 319]
Ultimately, large numbers of variants with small effects are likely to be important for athletic performance. This is consistent with other complex polygenic traits and makes the methodological challenges above difficult to overcome. Increased availability of whole genome sequence data on large numbers of individuals alongside athletic performance data would help to overcome these challenges. This would likely lead to better identification of genomic variation related to sporting ability.
Regulation
The regulations surrounding sporting competition are largely overseen by independent international governing bodies. These governing bodies are largely beyond the legislative remit of national governments.[footnote 320] Given the current infancy of genomic science for sport there is little to no guidance on genomics from the governing bodies involved.[footnote 321] As athletes are selected based on ability, the suitability of non-sporting genomics legislation (which mostly seeks to limit discrimination based on genomic factors) for regulating sporting activity is unclear.
An exception to this trend is the World Anti-Doping Agency (WADA), who have expressly prohibited gene doping.[footnote 322] WADA are considering the rollout of genomic sequencing to monitor athletes’ genomes for any changes as part of the Athlete Biological Passport framework.[footnote 323] WADA published its first laboratory guidelines in January 2021 to directly detect gene doping using polymerase chain reaction (PCR) based techniques.[footnote 324]
With regard to eligibility, World Athletics endorses genetic testing for sex verification, but challenges to this are often pursued as a competitive sporting issue and not as a legal discrimination issue. This can be seen in the cases of Dutee Chand[footnote 325] and Caster Semenya.[footnote 326] In the near future there may be risks of gene doping that is undetectable by today’s standards, such as through the potential use of RNAs.[footnote 327]
The only other limitations on genetic testing which are imposed in this area are the general legal regulations pertinent to the use of the technology. This relates to an individual’s right to privacy, to use of their data, and to the way that genomic information may be integrated with other protected characteristics in an employment context as discussed above in the Employment section.
Ethics
The use of genomic information in sport raises several ethical questions. First, the identification of athletic potential via a genetic test may result in an individual being coerced into pursuing an activity or programme. If the athletic aptitude does not manifest, or the individual’s performance fails to match expectations, this could have a negative effect on their mental health. This is especially relevant where a parent or guardian is making choices on behalf of a minor, potentially limiting the child’s “right to an open future”.[footnote 328]
A test that fails to identify high athletic potential may nonetheless be irrelevant to an individual’s actual sporting ability. The current accuracy of genomic prediction for many characteristics relevant to sporting ability is low, making the prediction a poor guide to an individual’s likelihood of sporting success. A ‘negative’ test could discourage individuals from pursuing sporting activities that could contribute to their health and wellbeing. Such tests could also lead to stigmatisation and negative discrimination.
If genomic predisposition to injury could be identified, this could allow preventive measures to be taken to reduce potential harm. It could also cause them to cease the activity, to the detriment of their health and wellbeing. In the case of a professional athlete, the test could result in loss of livelihood, even without the condition manifesting. Justifying such action would be especially difficult if the test is of low accuracy or only shows a small increase in risk.
The case of professional basketball player Eddy Curry Jr illustrates some of these issues. His employer, the Chicago Bulls, sought to compel him to undertake a DNA test to determine his susceptibility to a heart condition (hypertrophic cardiomyopathy, which can lead to sudden cardiac arrest).[footnote 329] Curry refused to take the test and challenged it as an infringement of his right to privacy. He was traded to another team before the issue was resolved, and his new employers did not demand a DNA test.
Academic specialists generally suggest that in balancing the different interests at stake, athletic programmes should aim to facilitate player success, promote player safety, and avoid genetic discrimination within and beyond the programme.[footnote 330] A 2019 study of regulation and genetic testing in sport[footnote 321] concluded that:
The tendency of the law to treat discrimination in sport differently to other areas of society could leave athletes vulnerable. Whilst genetic information may be useful for understanding genetic traits and their relationship with athletic performance, going beyond this to select athletes on the basis of genetics is discouraged and the interests of sport should be fairly balanced against the human rights of the athlete”.
Other ethical issues may arise from the competing imperatives of the professional athlete. In theory a gene variant that provides an athletic advantage could also increase susceptibility to certain diseases or ill health. A possible example is Malignant Hyperthermia Susceptibility (MHS), the causes of which have a genetic component, and which has been associated with muscular individuals and elite athletes.[footnote 331] This would further complicate the ethical questions around testing, selection and participation.
Education
Education is one of the most heavily studied non-medical traits in terms of genomics. It was the focus of the first large-scale GWAS of a social-science phenotype published in 2013.[footnote 332] Much of the focus on genomics in education builds upon the concept of personalised education; tailoring an individual’s educational environment and experience to support their learning in the most effective way.[footnote 333]
Most genomic education research has focused on educational attainment, defined as the number of years of education that an individual has attained. A smaller amount of genomic research has been conducted on educational achievement, defined as an individual’s performance at a given level of education. This is because data on educational attainment is more widely available than data on educational achievement. For example, the largest GWAS of educational attainment to date was able to use a sample of 1.1 million participants, compared to a sample of 565,000 for self-reported mathematical ability.[footnote 22] This may be for several reasons, including that attainment is easier and quicker to collect than achievement; there may be less recall bias in attainment than achievement; and attainment is more easily standardised internationally than achievement.
There is also a large body of genetic research into cognitive phenotypes such as intelligence, which relates strongly to education.
Key messages and considerations for policy
-
Educational attainment and achievement are highly polygenic traits, with heritability estimated between 40 percent and 60 percent by twin studies.
-
It is difficult to accurately predict a given pupil’s educational achievement from their genome using currently available polygenic scores.
-
Genomic screening could in future provide an opportunity to target educational interventions at those with learning disabilities much earlier than would otherwise be possible.
-
The strong role of environmental factors in education makes a purely genomic approach difficult. Isolating the genomic component of educational attainment may however allow fairer creation and assessment of learning environments.
-
Testing children for learning disabilities or variations in educational performance, personality and cognition raises ethical challenges. For example, does the potential benefit of identifying pupils in need of additional educational support outweigh the cost of stigmatising these pupils?
-
Current genomic knowledge is heavily weighted to populations of European ancestry. This means that the potential benefits of genomic knowledge and applications are likely to apply unevenly to UK citizens, being most strongly supportive of already advantaged groups.
Genomics in education today
For testing, assessment and streaming, there is currently no use of genomic testing or genomic information within the UK education system. There have not yet been any public statements or policy on genomics and education by the UK government.
There is increasing academic debate on the potential role of genomics in education, particularly for identifying pupils who may be most in need of further or specialist academic support.[footnote 334] While genomics is not being applied within education in the UK at present, a huge amount of research has been undertaken to understand the genomics of education and its potential applicability. Key areas of research and current knowledge are discussed below.
Education and intelligence
Twin studies have estimated the heritability of educational attainment at around 40 percent[footnote 335]. The largest GWAS to date identified 1,271 SNPs that were strongly associated with educational attainment. In a polygenic score these SNPs explained 11-13 percent of the variance amongst individuals of European ancestry, far short of the estimated twin heritability[footnote 22]. This “missing heritability” gap demonstrates that educational attainment is a highly polygenic trait with many relevant SNPs still to be identified. Genomic prediction of attainment is similar to that provided by some environmental measures such as household income but outperformed by others such as parental education and family socioeconomic position.[footnote 22][footnote 336]
Twin study heritability estimates for educational achievement are around 60 percent[footnote 337][footnote 338]. The largest GWAS to date identified 618 SNPs strongly associated with educational achievement (defined as self-reported mathematical ability), which together only explained 3 to 5 percent of the variance in high school grade point averages[footnote 22]. The missing heritability gap for achievement is therefore currently around 55 percent.
Twin studies have estimated the heritability of intelligence as captured by performance on IQ tests at around 50 percent,[footnote 45] though this increases with age from around 20 percent in childhood to 80 percent in adulthood.[footnote 44][footnote 45] 336 SNPs have been identified for IQ test performance which together explain 5 to 7 percent of the variation in cognitive test performance, making the missing heritability around 45 percent.[footnote 22],[footnote 339]
Each of educational attainment, educational achievement and cognition have been demonstrated to be highly polygenic traits.[footnote 22][footnote 340] That is, they are characterised by many SNPs with small effect sizes, each accounting for only around 0.01 percent of trait variation.[footnote 22] Therefore, there is no evidence for substantial effects on education from a single genetic marker. The missing heritability gap is likely to be reduced with larger sample sizes and the use of whole genome sequencing, which will improve the identification of smaller effect sizes and rare variants respectively.
Identifying the biological basis of education and cognition
GWAS into education and related phenotypes have provided insight into the biological architecture of learning and education processes. For example, genes previously implicated by GWAS are involved in brain development and neuron-neuron communication and are highly enriched for expression in the hippocampus and cerebral cortex.[footnote 22]
Future genomic studies may help to further uncover the biological basis of learning and cognition. This may help to better understand how pupil’s education can be more effectively supported, enable earlier identification of individuals in need of support, and inform more beneficial interventions to improve pupil’s educational outcomes.
Predicting student achievement from genomics
On average, pupils with higher polygenic scores for educational attainment achieve higher grades than those with lower polygenic scores, both generally and within specific subjects such as mathematics.[footnote 22],[footnote 341] While this is broadly true, it is nevertheless very difficult to accurately predict a given pupil’s educational achievement using currently available polygenic scores.[footnote 342]
Education is influenced by a complex interplay of genomic and environmental factors. While these factors in combination may offer the best prediction for educational outcomes,[footnote 336] it is questionable how much information current polygenic scores add to prediction accuracy above already available or more easily collected social/environmental data. A recent study found that while genomic information was broadly as predictive of educational performance at age 16 as measures of socioeconomic background, it provided no improvement above measures of prior achievement.[footnote 5] While polygenic scores will become more powerful in the future as genomic research advances, it is uncertain if, and when, they will be sufficiently accurate for predicting a child’s education.
However, there is very little stopping DTC genomic testing providers expanding into education-relevant fields and marketing these tests to parents. Three DTC genomic testing providers were offering genetics-informed IQ tests from a saliva sample in 2018.[footnote 15] This was ahead of the science, with the predictive power of polygenic scores for intelligence at the time only explaining 1 percent of the variance in intelligence. This has subsequently improved, but remains fairly low at 7 percent.[footnote 343] The utility and ethics of such tests (especially during embryonic testing) is heavily disputed.[footnote 344][footnote 345]
As discussed in Chapter 4, there is little regulation of the DTC genomic testing sector. If these tests gain traction and see widespread use in the consumer market, schools and parents are going to need support to navigate the issues that these predictions raise.
Early identification and interventions
Genomic data can be measured at birth, prior to other data used by educators being available.[footnote 55] This means that it may offer earlier and more targeted personalisation or interventions that are designed to improve educational outcomes. This could for example consist of identifying students in need of academic support, designing approaches to learning, or targeting treatments for pupils with learning disabilities.
Accurate genetic screening for such disabilities is not yet available[footnote 344][footnote 346], but is likely to improve in the future, offering transformative opportunities for education years before a formal diagnosis would otherwise be possible.[footnote 344] However, it will be vital that false positives and negatives are considered when screening for learning disabilities to ensure no harm to those incorrectly identified as a result of screening.
Looking to the future: what else could genomics tell us?
Education is highly dependent on the environment and improved genomic knowledge may help to better understand how environmental factors affect education. By isolating genetic effects on education, researchers may be able to more accurately determine how the environment affects pupil performance. Environmental factors here may include schools, teachers, and teaching methods.
A further strength of genomics in this area may lie in investigating differences between groups of individuals. For example, research has demonstrated genomic differences between pupils in different schools[footnote 347][footnote 348] and streams. These genomic differences may underlie measures of class and school performance, unfairly benefitting or disadvantaging institutions or teachers on school league tables or performance metrics.
There is also evidence for gene-environment interactions in some situations whereby genetic and environmental factors combine with each other to influence education. A recent study found that students with higher polygenic scores for educational attainment were more likely to enrol in and complete advanced mathematics courses than students with lower polygenic scores, regardless of school type. The study also found though that students with lower polygenic scores were less likely to drop out if they were enrolled in more economically advantaged schools. Another study found a similar pattern for enrolment in higher education; the college enrolment gap between students with higher and lower polygenic scores was smaller amongst pupils from higher than lower status schools.
However, there are reasons to treat genomic research of education with caution. Different groups of individuals experience systematically different (unequal) environments, and it may therefore be difficult to accurately disentangle whether differences in genotype cause differences in education. Genomic studies could therefore be incorrectly attributing environmental effects to the genome.[footnote 65] Education is highly contextualised and educational differences between individuals may be larger for environmental than genetic factors.
The degree to which genotypic differences between individuals may result in differential educational performance will likely rely on the way in which education systems are designed.[footnote 349] For example, over the course of the 20th Century there were huge changes in norms and attitudes relating to the education of girls in many Western countries. Had a genomic analysis been conducted 100 years ago, genes on the Y chromosome may have associated with educational achievement. This clearly would have reflected a social rather than genomic mechanism, even though it would have been seen in genomic data.
Regulation
Clearly there are several challenging technical and ethical considerations relating to the use of genomics in education.
However, there is currently no regulation in the UK relating to the use of genomics in education or genetic discrimination. The EU Charter of Fundamental Rights (Article 21.1) prohibits discrimination based on genetic features under EU law. The UK Equality Act 2010 prohibits discrimination and harassment related to certain personal characteristics, though this does not explicitly include genomics.
The lack of regulation explicitly on the use of genomic data in education contrasts with some other nations. Three states in the USA (California, West Virginia and Washington) have implemented genetic non-discrimination protections relating to education. These laws prohibit discrimination based on genotype by schools (California), the sharing of confidential student information including genotype data (West Virginia), and the collection of biometric data by state agencies (Washington).[footnote 350]
Ethics
There are numerous ethical challenges surrounding the use of genomic information in education:
-
Ancestry portability. Current genomic knowledge obtained from GWAS are restricted to individuals of European ancestry. While a polygenic score from the latest GWAS explained 11 to 13 percent of the variance in educational attainment amongst individuals of European ancestry, it explained only 1.6 percent of the variation amongst an African American sample.[footnote 22] If this polygenic score was used to identify students in greater need of educational support, it would more accurately do so for (and therefore disproportionately benefit) students of European than African American ancestry. This could result in the reinforcement and further development of social inequalities in education.
-
Choice. Testing could potentially help individuals to make more informed choices about their education with the view to improving their outcomes. However, it could also reduce autonomy if the person feels compelled to take the test. If a parent chooses for a child to be tested, this can affect the child’s “right to an open future”. Adverse results could also lead to fatalism or stigmatisation and discrimination (the Golem effect).
-
Selection. Broad genotypic differences in the intakes of schools and streams within schools have been demonstrated.[footnote 349] It is possible that schools could use genomic tests to justify the (lack of) enrolment of pupils based upon their perceived educational potential. Similarly, it is possible that polygenic scores could be used to ‘stream’ children into routes they might not choose or otherwise follow. Given the inability of current polygenic scores to accurately predict performance and the Eurocentric focus of GWAS, such selection decisions could unfairly discriminate against some groups of pupils.
-
Misunderstanding. It has been argued that stigma and a lack of genomic literacy amongst educators could raise the risk of genomic data being misused in educational settings.[footnote 338] If pupils are enrolled or streamed by genomic factors, there may be deleterious mental health consequences for those that are perceived as less academically capable.
-
Non-pathological conditions. Testing for pathological phenotypes that have deleterious effects on education could allow effective preventive or ameliorative interventions to be deployed. However, testing for non-pathological phenotypes such as cognition or personality raises ethical concerns because these traits do not harm individuals, and their value is socially determined. Furthermore, genomic screening for complex traits are predictive, not deterministic, and the benefits must therefore be considered against risks such as stigmatisation. As a result, the Nuffield Council for Bioethics suggested caution about testing children for phenotypes such as these.
Criminal justice
As we have seen in Chapter 4, forensics has utilised genetic technologies for some time, and there is a large amount of research occurring at the intersection of genomics and criminal behaviour, with a range of complex ethical implications. Whilst genomic information is not currently utilised within the UK’s criminal justice sector (and there are no plans to do so), genomics has been used in a small number of cases within the justice systems of a few other countries, to shape verdicts and influence sentencing. It is plausible that people may seek to use genomics in the future for behaviour prediction. As part of our horizon scanning and consideration of what might be possible, here we explore some of the background to, and wider interest in, the role that genomics could play within criminal justice.
Key messages and considerations for policy
-
There are correlations between certain genes and characteristics that can influence criminal behaviour. Alleles of the MAOA (Monoamine Oxidase A) gene have been linked to aggressive behaviour. Substance Abuse Disorders (SUD) have been linked to specific genes, with a tendency to abuse substances having high heritability.
-
Polygenic scores which are indicative of a tendency towards criminal behaviour have not been identified, however polygenic scores for disinhibition may serve as a proxy measure.
-
Genomic evidence has only been submitted in a small handful of cases internationally as a mitigating factor (though none of these cases have occurred in the UK). The use of genomic data in this way may become more frequent in some countries as the evidence base improves.
-
The ethical implications of using genomics to predict criminal behaviour are particularly fraught, raising questions around choice, autonomy, consent, avoidance of harm, equality, fairness, and inclusion.
Genomic influences on criminal behaviour
One of the first connections made between criminal behaviour and genetics provides a cautionary tale. Klinefelter syndrome affects approximately 1 in 1000 men, and is associated with infertility, cognitive impairment, low testosterone, reduced education and higher rates of unemployment. Studies from the US and UK in the 1960s and ‘70s[footnote 351][footnote 352] claimed that people diagnosed with the syndrome committed more crime, especially with regards to sexually-motivated crimes. However, subsequent research has cast doubt on the association between Klinefelter syndrome and crime due to poor study design, prejudice, and ascertainment bias in the original studies.[footnote 353] Recent studies have posited that in fact, the poor socio-economic conditions of men with Klinefelter syndrome may be a larger influence on their patterns of offending than any genetic predisposition.[footnote 354] The complexity of influences on behaviour and resulting criminal activity must not be underestimated.
Nevertheless, many studies have since sought to further identify genomic influences on criminal behaviour, with mixed results. Twin studies across several nationalities have suggested that severe anti-social behaviour (ASB) has a strong genetic component[footnote 355] and that there may be some common genetic factors between ASB and other cognitive and psychiatric traits. The Broad Antisocial Behaviour Consortium published findings of the largest GWAS on European individuals for ASB, in which they were unable to find any specific genetic variation related to ASB.[footnote 356] They did however emphasise that the study was limited in sample size and that there was a lack of standardisation on the ASB measure across the samples used. Future studies with greater statistical power may lead to the possibility of polygenic scoring to infer individual ASB risk in the future.
A small number of genes are correlated with criminal behaviour, particularly violence. A recent Finnish study has implicated MAOA variants as well as variants in the CDH13 gene[footnote 357] as being involved in extremely violent behaviour.[footnote 358] The study investigated the genotypes of over 500 violent criminals, all of whom were defined as receiving a prison sentence for at least one homicide, attempted homicide, or battery. Participants were specifically genotyped for MAOA and CDH13, which revealed a strong correlation between variants of these two genes and extremely violent behaviour (defined as less than or equal to 10 violent crimes). From these findings, the study authors conclude that approximately 5 to 10 percent of all severe violent crime in Finland could be attributable to these specific MAOA and CDH13 genotypes alone. The study emphasised that the sensitivity and specificity of the genotype findings are too low for a formal screening programme. However, as a precaution it suggested that criminals with this genotype should be encouraged to avoid substances such as alcohol or amphetamines that cause transient dopamine bursts, due to their role in aggression.
Whilst genes or polygenic scores indicative of criminal behaviours are yet to be established, polygenic scores for other related metrics may act as a proxy indicator. A longitudinal study[footnote 359] of British and New Zealander participants in 2018 found that individuals with lower polygenic risk scores for educational attainment were slightly more likely to commit antisocial behaviour/have a criminal record. Another study of the same cohort showed a similar link between criminal behaviour and the age at which participants had their first child (age-at-first-birth, AFB)[footnote 360]. Researchers were able to derive a polygenic score to predict individual AFB, which was used as a metric of disinhibition: individuals who scored low exhibited poor self-control in childhood, were more likely to have a substance dependence, and were more likely to engage in criminal behaviour, with associations persisting when adjusted for age of puberty onset.
Genomic influences on substance abuse and addiction
Contrasting current knowledge of gene-influenced violent or antisocial behaviour, knowledge of the genomics underpinning substance abuse is much more established. Broad demographic twin studies have revealed that substance abuse has a strong genetic component. Cannabis addiction, alcohol dependence and cocaine use disorders have heritability estimates of 51 to 59 percent, 48 to 66 percent, and 42 to 79 percent respectively.[footnote 16] While substance abuse is not itself always a crime, the surrounding behaviours may well be, and is where the substance itself is illegal.
Furthermore, many GWAS have identified the genetic markers related to substance use disorders. These studies have identified numerous possible targets for pharmacological intervention. They suggest that increasingly powerful polygenic scores will be available for general and specific predisposition to Substance Use Disorder (SUD)[footnote 361] as larger cohorts of WGS become available. Generally, scientific consensus[footnote 362][footnote 363] agrees on the following:
-
SUDs are highly polygenic with multiple genes of small effect contributing to SUD risk, much like behaviours discussed earlier in this chapter.
-
A large fraction of the genetic risk for SUD is not specific to one substance; rather, the genetic risk is general to different types of substances with specificity being conferred by the environment. This may indicate that there are common pathways (for example, via dopaminergic neurotransmission) connecting problem use of multiple drugs.
-
Many genetic associations with SUD are also shared with psychiatric disorders including externalising disorders such as impulsivity and disinhibition.[footnote 364]
-
Genes associated with specific disorders have also been identified. Genes have been found with an association to smoking (accounting for 10.9 percent of phenotypic variation in UK cohorts)[footnote 365] and misuse of drugs: alcohol (with a heritability of 12 percent in a UK Biobank/23andMe cohort)[footnote 366], cocaine (around 65 percent heritability in a US cohort)[footnote 367], heroin (22 percent heritability in a Han Chinese cohort) and methamphetamines (18 percent heritability, in a Han Chinese cohort)[footnote 368].
-
Genetic background may also be important as not all results are replicable across ethnicities – for example a variant strongly associated with opioid addiction in African Americans is not always associated in European Americans.
-
Polygenic scores for substance abuse are a real possibility: for example, polygenic scores for alcohol abuse (involving 11 risk loci) can account for 12 percent of all positive alcohol use disorders identification test (AUDIT) scores, in UK Biobank/23andMe cohorts.[footnote 369]
Genomics in criminal justice today
Genomic analysis has been introduced as evidence in only a small number of court cases around the world (none of which have been in the UK). Variants of the MAOA gene have been implicated in the development of aggressive and antisocial behaviours. A 2017 study found that genomic analysis of individuals possessing low-activity MAOA variants has been submitted as evidence in 11 criminal cases worldwide.[footnote 370] The precise effect that genomic evidence had on these results is unclear; some sentences were lowered but the reasons for this were not provided. Genotypic evidence may not be persuasive to courts because the precise impact of the allele on individuals is difficult to establish. This, combined with the infrequent use of genomic evidence, has led researchers to conclude that genomic evidence has had little systematic impact to date on judgments or sentencing.[footnote 371]
Looking to the future: what else could genomics tell us?
Criminal and anti-social behaviours are complex traits, with studies indicating that they are shaped by a range of environmental and genomic factors. Should the evidence indicate that the use of genomics in criminal justice is feasible, fair and scientifically sound, it is plausible that in the future people may seek to utilise genomics to:
-
Estimate an individual’s genetic predisposition towards certain traits, to explain or mitigate some criminal behaviours as part of a criminal case, potentially seeking to influence sentencing.
-
Identify individuals who are genetically predisposed to behaviours associated with criminal behaviour, such as substance abuse.
In the future, testing could, in theory, allow personalisation of interventions to deter people from crime or to improve rehabilitation outcomes. Genomic studies could also help identify the biochemical causes of behaviours behind many of these traits. This could potentially help to identify targets for pharmacological interventions.
Technical limitations
Polygenic scores currently provide poor prediction at the individual level, meaning that they may mischaracterise the observed trait for a given person. They also have limited predictive accuracy in samples where the subject differs in ancestry or environment from the GWAS discovery sample[footnote 372]. Polygenic scores also only provide a measure of genomic predisposition, meaning that the predicted trait may not manifest.
Genomic prediction doesn’t assess environmental influences, which may be equally as important in influencing criminal behaviour. So, whilst the predictive power of polygenic scores is likely to improve with time, other predictive policing approaches may be more effective, which could include the use of proxy indicators. These can be place-based (looking at places and times where crime is more likely to occur)[footnote 373] or person-based (looking at socio-economic factors that make a person more likely to be a perpetrator or victim of crime, or prior incidences of offending).[footnote 374] Genomic information could theoretically be used to augment these approaches. However, concerns about the ethics and transparency of predictive policing approaches[footnote 375] would be just as applicable to using genomic information for this purpose.
Ethics
To date, the use of genomics within the criminal justice system is virtually non-existent globally, and is certainly not utilised in the UK. However, it is plausible that there may be general interest in the role genomics could play, even if only at a theoretical level. We do know that some traits relevant to criminal justice are heritable, and genes associated with violence, proxy indicators for likelihood to engage in criminal behaviour, and substance abuse have been identified. Genomic evidence has been submitted as mitigation at trial in a small number of cases (though none in the UK) and this may become of interest more widely in future. Whether or not this is possible, advisable or desirable, it may be worth considering at a hypothetical level as part of horizon scanning and future-proofing.
As in other fields, the potential use of genomic information in the criminal justice system raises complex ethical issues in terms of choice, autonomy, consent, avoidance of harm, equality, fairness, and inclusion. These issues go beyond the scope of this chapter but will be considered briefly here.
Genomic testing for the purposes of criminal justice would be fraught with issues. The circumstances of the test would be critical: who might be chosen for testing – or volunteer for testing – and why? Would the individual freely consent to a test, be coerced, or could a parent choose to have their child tested? Other questions include:
-
Would it be acceptable to offer genomic testing that enabled preventative intervention but could also cause harm or distress for the individual and family members, given the shared basis of DNA?
-
Would it be fair to offer genetic testing to individuals to potentially mitigate/reduce a sentence, and would this be more or less fair than current approaches?
-
Would it risk stigmatising individuals as “genetically criminal” based on a prediction of a predisposition that might never result in a crime?
-
Would a genomic diagnosis of a predisposition to criminality become a “self-fulfilling prophecy”, with the individual feeling that such behaviours are inevitable and beyond their control?
The concepts of acceptable accuracy and fairness are also relevant. A decision may be fair to all people in principle, but if that decision is based on data that is biased against a particular population group, it could be considered discriminatory and unfair. A relevant example could include using a hypothetical polygenic score to assess the risk of violent reoffending - this could be considered discriminatory if the test results were found to be more accurate for some groups than others. Indeed, as discussed earlier, current genomic datasets are notoriously biased due to their selectivity for individuals of European ancestry, making this a real possibility. This potential for bias will be reduced by increased use of whole genome sequencing and efforts to increase the diversity of genomic databases.
Insurance
Genetic tests, and a better understanding of how genomics affect health and behaviour, could improve how insurance companies assess eligibility for cover and set premiums. However, there are issues of ethics and fairness associated with the use of genomics for insurance, and regulation on this issue varies around the world.
Key messages and considerations for policy
-
In the UK, there are strict limitations on the use of health-related genomic information for insurance purposes. This is set out by a voluntary code agreed between the government and the insurance industry.
-
Polygenic scores for heritable behavioural characteristics such as risk-taking behaviour, and physiological factors such as susceptibility to injury, could inform insurance policies in future.
-
Direct-to-consumer (DTC) genomic tests could increase information asymmetry between the insurer and the insured, leading to increased costs, though to what extent is unclear.
-
Improvements in the accuracy and specificity of polygenic scores could make them more useful for insurance, however these uses would have to overcome public resistance, and would need to satisfy the requirements set out in the Concordat and Moratorium on Genetics and Insurance.
Genomics in insurance today
The UK insurance industry currently follows a voluntary code setting strict limitations on the use of health-related genomic information in determining eligibility for insurance.[footnote 376] The potential use of genomic databases (like the UK Biobank) by commercial entities is regarded as posing some ethical dilemmas.[footnote 377] For the UK Biobank in particular, its ethics and governance framework acknowledges that commercial entities may wish to access the data.[footnote 166] However, that access is only granted for uses that support the purpose of the UK Biobank, which likely excludes insurance companies. Meanwhile, insurers already use risk proxies (for example, personal and family history, demographics) to calculate premiums, leading to higher costs for some groups (for example, car insurance is more expensive for young, male drivers). Under this approach many risk factors are unknown, uneven risks are pooled and low-risk individuals in effect subsidise those with higher risk. Improved genomic information does not necessarily have to change this fundamental dynamic.
The public has concerns about adverse selection by insurance companies. What is considered actuarial fairness and best practice (for example, using personal and family history, and pooling risk evenly across a number of contributors) may not always match what the public considers to be social fairness. Research commissioned in 2019[footnote 378] involving a public dialogue on genomics suggests insurance is an area of distrust, and there are concerns about inappropriate surveillance or penalisation of people with acute healthcare needs.
Genetic traits relevant to insurance
The debate around using genomics for insurance has focused on health, considering how genetic evidence can improve understanding of an individual’s risk of disease or disability, and how this should be considered in underwriting insurance and claims. However, polygenic scores for heritable behavioural characteristics (for example, predisposition to risk-taking) may be just as relevant for insurance.
Risk-taking behaviour
Twin studies show that risk-taking behaviour is moderately heritable at around 30 percent,[footnote 379] though predictive estimates vary across studies. A GWAS of nearly 1 million people identified 124 variants across 99 separate genes that were linked to general risk tolerance. The study looked at both general propensity for risk (“would you describe yourself as someone who takes risks?”) and four risky behaviours (adherence to speed limits, alcohol consumption, smoking, and sexual partners).
46 of the 99 general-risk-tolerance loci were also associated with one of the four risky behaviours, suggesting an overlap in the genetics of these traits. Despite the large number of significant variants found, a polygenic score based on the GWAS had a predictive power of only 1.6 percent for people’s general risk tolerance.
Susceptibility to injury
As noted in the employment case study, twin studies suggest a genetic component to people’s susceptibility to injury; Salminen[footnote 286] found genetic influences account for around a quarter of the variance in injury involvement of 33 to 60-year-olds, whilst the environment was a stronger influence on younger age groups. A systematic review of the genetic factors involved in tendon and ligament injuries identified no single genetic cause, but associations with connective tissue component genes.[footnote 380] Specific injury types have also been found to have a high genetic contribution, with a twin study in 2020 determining that anterior cruciate ligament (ACL) rupture (a musculoskeletal injury common to contact sports and military service) has a heritability of 69 percent.[footnote 381]
Looking to the future: what else could genomics tell us?
Direct-to-consumer (DTC) genetic testing may exacerbate information asymmetry, where the insured have more information about their personal risk than the insurer. This could lead to “adverse selection”,[footnote 382] where genetic tests provide the applicant with knowledge of an increased risk which they are not required to disclose to the insurer. The applicant could then take out cover that does not price in their risk, potentially leading to a loss for the insurer and higher premiums for other customers.[footnote 383] Where employers provide health insurance there could be higher costs for businesses. However, if disclosure of genetic tests to insurers were mandatory, people may be deterred from taking genetic tests that might provide them with useful information on potential health risks.
The impact of unknown genetic risks on insurers is not clear. A study from 2011 estimated the impact of adverse selection on insurance premiums to be an increase of between 1 percent and 3 percent if both genetic tests and family history underwriting were disallowed, dropping as low as 0.1 percent of premium income if family history underwriting were permitted.[footnote 384] Other research has predicted that critical illness claims could increase by an average of 26 percent if the use of genetic information in underwriting is not permitted.[footnote 17]
Polygenic scores could lead to a greater understanding of personal risk, allowing more accurate pricing of insurance. Polygenic scores consider the total risk of developing a disease conferred by common (but individually low risk) gene variants, giving better predictive powers for polygenic diseases. They could reduce costs for those with lower polygenic risk but potentially raise them for those with higher risks. Insurers could also offer incentives for risk reduction, such as monitoring devices to encourage safe driving. A concern is that insurers could refuse to insure very high-risk individuals, or that the premiums for these individuals could become so high that they choose not to have insurance. This could lead to higher costs for wider society, for example if young male drivers who were unable to obtain affordable insurance chose to drive without insurance and were subsequently involved in accidents.
The extent to which DTC tests offer an accurate assessment of risk is also important. Testing for monogenic traits such as Huntington’s disease and cystic fibrosis is highly informative as genetic predisposition for these conditions conveys a high risk (see Chapter 3). Testing for polygenic traits is likely to be far more limited and predictions far less informative. This is evidenced for behavioural characteristics where even the most informative polygenic scores are able to predict only a fraction of total variance. Individuals may regard the results of low accuracy polygenic tests as more informative and deterministic than the science suggests. This could result in people under- or over-insuring if they perceive themselves as having high or low genetic risk.
Regulation
Countries vary in their approach to regulating the use of genetic tests for health insurance. A 2017 report by industry thinktank The Geneva Association[footnote 385] groups regulatory approaches into different categories (see Table 3, below). The report notes that most insurance companies have failed to keep up with the pace of change in genomics, particularly the possibilities offered by whole genome sequencing. Insurance companies also struggle with legal definitions of genetic tests.
Table 3: Different regulatory approaches to the use of genetics in insurance. Adapted from The Geneva Association.[footnote 386]
| Category | Description | Example countries |
|---|---|---|
| 1 | No regulation | India, China, Finland, Spain. |
| 2 | No regulation but does have codes of conduct from insurance industry groups | Greece, Japan |
| 3 | Prohibition for some types of insurance The Genetic Information Non-discrimination Act (GINA) makes it illegal for health insurance providers to use or require genetic information to make decisions about a person’s (employment and health) insurance eligibility or coverage but not life, disability or long-term care. Most US states have their own regulations, mainly relating to health | USA |
| 4 | Prohibitions on insurers requiring applicants to take a genetic test and prohibitions on discrimination if the applicant refuses to take a test | Australia |
| 5 | Prohibitions or moratoriums on using results from existing tests when policies are below certain limits | UK, Germany, Netherlands, Switzerland |
| 6 | Prohibition or moratoriums on using results from existing tests at all, sometimes including use of family history | Belgium, Austria, Canada, Denmark, France, Ireland Poland, Portugal, Singapore |
The UK approach: Concordat and Moratorium on Genetics and Insurance
In 1999, the UK government established the Genetics and Insurance Committee (GAIC), which approved an application to use Huntington’s disease test results for life insurance in 2001. This decision provoked an adverse reaction from the public and Parliament, including the publication of a critical report by the House of Commons Science and Technology committee.[footnote 387]
In response, there was a Moratorium in 2001 to 2002 that later expanded into the voluntary “Concordat and Moratorium on Genetics and Insurance”, which was agreed between government and insurance companies in 2005.[footnote 388] The concordat’s two core principles committed insurers to not asking customers about predictive genetic test results when applying for insurance (except in very limited circumstances) or requiring an applicant to undertake any form of genetic test to obtain insurance.[footnote 389]
The Concordat and Moratorium was replaced by the Code on Genetics and Insurance in 2018. It sets out principles by which insurers will operate and confirms that insurers will never ask for genetic test results for policies worth less than £500,000. Above that figure only Huntington’s Disease test results may be requested. The Code is open-ended, and will be reviewed every three years, allowing for the agreement to reflect developments in genomics. Insurers will not ask for the result of any predictive genetic test obtained through scientific research.[footnote 376]
The Association of British Insurers (ABI) reports on members’ compliance with the code, however not all insurers are members of the ABI, and it is possible for insurers to leave and re-join the ABI. The code also fails to specify any sanctions for insurers that fail to follow the code, though consumers can use this as the basis of a complaint to the Financial Conduct Authority (FCA).
Ethics
Using genetic tests to assess eligibility or set pricing for insurance policies has important ethical implications. There is a risk that people with heritable conditions, or whose genomic tests indicate they are at high risk of adverse outcomes, may be priced out of the market. Without strong governance and safeguards, the perceived risk of insurers accessing genomic data may in future affect public willingness to participate in research, consent to otherwise potentially beneficial screening or tests, or even take out insurance policies in the first place.[footnote 390]
Biases in genetic data would be a problem in their use for insurance purposes. The European bias of GWAS studies (noted in Chapter 3) means that prediction of predisposition to certain medical conditions may not be accurate for minority populations. Using these incorrect assumptions of risk to assess eligibility for insurance or to set premiums would disadvantage both the insurer and the insured. Racial disparities in access to genetic testing and subsequent healthcare interventions could also exacerbate these effects.[footnote 391]
Future advances in genetic testing may pose further dilemmas with regards to insurance. The UK code on genetic testing and insurance includes criteria that would justify insurers requiring that the result of a novel genetic test be disclosed in the future, but these criteria are framed around health issues rather than predisposition to certain behaviours. The criteria stipulate that:
-
The condition must be clearly and measurably heritable; with a high probability that those with the gene variant will develop the condition with a resulting materially increased likelihood of significant morbidity and mortality.
-
A predictive genetic test of high analytical and clinical validity must exist, and provide helpful clinical information about diagnosis, treatment, management or prevention of disease.
-
The insurer must also demonstrate that not addressing the condition would adversely impact other consumers and insurers.[footnote 376]
Polygenic scores for non-health traits such as impulsive behaviour are unlikely to meet these thresholds in the near term. However, as whole genome sequencing becomes more widespread, polygenic scores for relevant traits will become more powerful. Therefore, there may be increased pressure to include more genetic screening results in insurance applications, particularly for rare variants which have large effects on traits. However, clinical or screening services may be unlikely to provide genetic tests if no relevant interventions are available.
Genome modification beyond health
Genome modification beyond health
Our case studies spanning Chapters 4 and 5 illustrate the breadth of genomic influence over many traits beyond health. If the use of genome editing tools for traits in health were to become realised, there is a possibility that they may also be used to modify traits beyond health, as a means to enable human augmentation.
Such procedures could be for the purposes of improving a person’s soft skills, sporting ability, or to reduce genomic predisposition for criminal behaviour. There are even a few cases of this being considered or performed today,[footnote 392] albeit secretly, and ahead of what the science supports. This means that considering what this future could look like is important, even if this future may be several decades away.
Key messages
-
Genome modification may one day be used as a medical treatment for some genetic conditions – early proof-of-concept trials have shown it can cure rare types of anaemia in adult humans.
-
The modification of embryos comes with more risks – the first report of genome edited twins was met with intense opposition, the procedure didn’t work, and whether it had any harmful side effects is currently unknown.
-
If genome modification was to become commonplace for treating diseases in the future, then the use of these same tools to modify non-health traits may also be a possibility.
-
The use of such tools in this way raises many ethical challenges, such as equity of access to these technologies, unfair competitive advantage, and entrenchment of inequalities.
Genome editing tools may help in curing human disease. In 2020 and 2021, two teams of researchers independently announced the first successful applications of CRISPR-Cas9 as a gene therapy. Patients with two different inherited types of anaemia[footnote 393][footnote 394] were cured by modifying progenitor red blood cells in vitro to express healthy forms of haemoglobin. Early phase I clinical trial results using CRISPR to genetically modify white blood cells to fight systemic cancer has also now shown to be durable and tolerable.[footnote 395] These cases demonstrate the potential of gene editing for therapeutic purposes. As our knowledge of the genomic influences of traits improve, so will our theoretical ability to influence them through genome editing.
But the prospect of using genome editing on humans comes with risks. Current genome modification methods sometimes come with off-target effects – this means that these tools may make unintended modifications to other genes in addition to the target gene. Furthermore, as discussed previously, many of our traits result from the influence of hundreds of genes in combination. Modifying one gene might therefore influence another trait for which a link is yet to be established. If these changes are made as germline mutations, these effects can also be carried across generations.
Recent reports of the first genome-edited twin girls show this to be the case.[footnote 396][footnote 397] The Chinese twins, born in 2018, were controversially modified as embryos to be resistant to HIV. However, genomic analysis of the twins after birth showed that the modification was incomplete and had therefore not worked. In addition, further research has shown that the intended genomic modification also effects cognition in mice, and in human brains following a stroke.[footnote 398] This could also potentially (and unpredictably) impact the girls’ academic attainment in the future,[footnote 399] or shorten their life expectancy.[footnote 400] These reports have been met with strong international condemnation and the researchers behind the experiment jailed and fined.[footnote 401]
But there are already a few cases of people considering or using genomic modification tools to modify their non-health traits. Examples of these include some professionals in sport (see box below), and a few high-profile cases of lone biohackers in the US. This is partly enabled by the fact that custom CRISPR solutions are freely available to buy online for educational purposes.[footnote 402] These solutions are then adapted for self-administration, in an effort to improve the individual’s health or physique.[footnote 403][footnote 404][footnote 405]
While there is little evidence that these interventions have so far produced any effect, the risk of side effects is considerably high given the lack of clinical/regulatory oversight. In response, the US Food & Drug Administration has outlawed the sale of DIY home gene editing kits marketed for human use within the US,[footnote 406] and WADA banned gene doping in 2018.
Gene doping is a non-therapeutic use of gene therapy used to improve performance in sporting events. Interest in gene doping began in the late 1990’s with the creation of a genetically modified mouse which over-expressed insulin-like growth factor 1, giving the mouse increased strength. Later modifications targeted the genes for erythropoietin and PPAR gamma, altering red blood cell capacity and fat metabolism, respectively. The scientists responsible for this research were contacted by coaches and athletes on the potential human applications of their work. This led WADA to ban gene doping in 2003, and gene editing in 2018, with athletes participating in the 2016 Rio Olympics now being retrospectively tested for additional doped copies of the EPO gene.[footnote 407][footnote 408] However, some are concerned that WADA lacks the technical capacity to detect these doping practices effectively and, so far, no positive cases have been found.[footnote 409]
The use of genomic modification as a route to human augmentation could have wider societal impacts. In 2021, the Development, Concepts and Doctrine Centre (part of the UK Ministry of Defence) released their report on human augmentation. In it, they state that genomic engineering has the most future potential as an augmentative technology, but that to pursue genomic modification across society risks entrenching social classes and leaving behind “an augmented underclass as relatively disadvantaged as the illiterate are in today’s societies”.[footnote 410] Furthermore, if countries were to explore the use of genomic engineering within their armed forces to create biologically enhanced soldiers, this would represent a significant asymmetric threat.
In the UK, the public’s opinion on human genome modification is nuanced. Figure 19 (overleaf) outlines the results of recent YouGov analysis which surveyed the acceptability of genome editing for several human-focused applications amongst UK adults. These uses ranged from clinical applications (to treat inherited diseases) to appearance modification. These data show that the acceptability of genome editing varies in part of its intended application, though the public is more positive about the use of genome editing for medical or clinical applications.
Figure 19: Public opinions on human genome editing for different applications since 2020. Source: YouGov.
Data, security, and public attitudes to genomics
Developments in genomics are changing how we conceptualise privacy and anonymity, with implications for data security in research and commercial databases. This chapter highlights some of the major issues surrounding data privacy and security and examines public attitudes to genetic testing.
Key messages:
-
Genomic data are a valuable resource requiring protection from inadvertent, unintentional, or unwanted disclosure. Direct-to-consumer genomic testing poses privacy risks which the public might not be fully aware of.
-
The privacy risks posed by genomic information are not limited to the individual to whom it belongs. Their immediate family and close relations may also be affected by any disclosure.
-
Genomic research projects (like the UK Biobank) are seeking to maximise the utility of large genomic datasets whilst minimising the risk to individual privacy. They use a variety of approaches, including mediated access to data through dedicated portals and data encryption schemes.
-
Best practice regarding the privacy and security measures employed by large genomic research projects in the UK should be regularly reviewed and understood by policymakers to ensure maximum data protection for UK citizens.
-
A purpose-built legal framework for genomic databases could provide clarity for the organisations that run the databases (whether commercial businesses or research bodies) and those who provide their genomic information.
-
The question of whether genes can be patented remains controversial. Companies contend that their genetic discoveries represent valuable intellectual property that should be protected, whilst others see this as limiting open research and leading to increased costs for medical tests.
-
Policymakers may wish to consider whether the current intellectual property system encourages innovation amongst biopharmaceutical companies, or if enhanced patent systems might limit research.
-
Public opinion is generally positive about the potential benefits of genomics, particularly for forensic science. However, they retain some ‘red-lines’ on issues where they feel genomics could disadvantage vulnerable people and are wary of private businesses accessing their genomic information.
-
Dialogue with the public about the current and future uses of genomics could allay their concerns.
Data privacy
The public benefits of genomics must be balanced against their potential impact on individual privacy. Large data collections and biobanks play an increasingly pivotal role in realising the public benefits to scientific research from genomics. Secure storage of data is a prerequisite, but research also requires that data are appropriately accessible and shareable to researchers within the terms of participant consent. This balance of value from genomic data and individual privacy has guided genomic research into health and will similarly underpin genomic research into non-health applications.
Participant involvement in studies may offer a valuable way of helping to ensure data privacy and security. Genomics England has a “Participant Panel” consisting of participants from the 100,000 Genomes Project.[footnote 411] The panel acts as an advisory body to the Genomics England Board, with the aim of ensuring that data held by Genomics England is used in the participant’s best interests.
Consumer-facing genetics services allow people to investigate their ancestry and genetic propensity to various traits, but also pose privacy risks. The largest direct-to-consumer (DTC) genotyping companies now hold data on millions of individuals. Because some companies allow parents to have their children tested, not all these individuals have provided informed consent. Some non-genomic information held on these individuals is also used for research (where consent is provided), but how this process is managed by the company can lead to an additional level of exposure.
There are a range of possible risks to privacy if genomic data are disclosed:
-
An individual’s genomic data contains personal information that they may wish to keep private. Disclosing this information would be unfair and could lead to stigmatisation or discrimination. For example, genomic information that suggests a high genetic predisposition to certain health traits could in principle be used by health insurance companies to deny cover or charge higher fees. Genomic data are also immutable: once disclosed, they will present a privacy risk for the remainder of that person’s life.
-
An individual’s genomic data may also partly reveal their relatives’ data. This could be through revealing relatives’ identities (see Chapter 4) or their genomic predisposition towards traits. It is unlikely that consent will have been sought from all relatives when an individual is genotyped. Therefore, relatives will likely have little control over this partial release of their data and potential intrusion into their privacy.
-
Stigmatisation of specific populations is a risk if genetic predispositions are perceived to be more prevalent among them than among other populations.
-
There is a risk to long-term research from these individual risks; if people fear that their genomic data will be disclosed or their confidentiality breached, they may be less likely to participate in research.
Risks to third parties can be reduced if companies are explicit that only first parties can submit samples for genotyping. This may consist of forbidding parents from submitting samples obtained from their children and restricting people from uploading genetic profiles that are not their own. Database holders could also limit multiple queries to prevent phishing attacks designed to extract large amounts of information.
There is a case for including such measures in codes of practice for the sector, however this is complicated by the UK’s genetic regulatory environment. There is “no purpose-designed legal framework or dedicated legal instrument” for genetic databases in the UK, which are instead governed by “a bewildering array of statutes, legislative provisions, regulations and common law doctrines, together with well over 30 codes of practice”.[footnote 412] This patchwork of regulation has led to inconsistencies in enforcement, with data breaches incurring only financial penalties. This compares to breaches involving tissue samples which can be punished by imprisonment. Breaches in tissue samples may be considered more serious than in genomic data, but both contain rich and personal information that is specific to a given individual. Gibbons[footnote 412] also stated that “creative, principled, […] practically workable and properly coordinated reforms are urgently needed” to facilitate the operation of genetic databases in the UK. There is still a lack of clarity in the law, though the UK’s flexible protections of statute, contract law and good governance may offer some protections. However, this may depend on the applications of GDPR and subsequent case law.
A recent report on DTC genomic testing from the House of Commons Science and Technology Committee’s inquiry into commercial genomics raised concerns from several contributors relating to data privacy.[footnote 111] The contributors questioned whether the consent that DTC genomic test customers provide to the use of their data is truly informed. They also highlighted the complexities around how genomic data might be used, and how advances in genomics and data science may complicate privacy in the future. The report advocated for the government to “review the adequacy of the UK’s data protection framework” for DTC genomic tests, and to advise customers on “the potential consequences of genomic test results for their relatives”.
The impacts of genomic data loss extend beyond the risks to the individual. Genomic data has significant value for research and commercial purposes, so accidental disclosure or theft represents an economic loss. An organisation from which genomic data has been stolen could be blackmailed for its return, or to prevent its release.
The US Federal Bureau of Investigation (FBI) has raised concerns about foreign access to the genomic data of US citizens, whether through legitimate scientific collaboration, funding of scientific research, or investment in and purchase of genomic sequencing companies[footnote 413]. This access could lead to national security risks through the potential identification of individuals. It could also lead to asymmetric access where the benefits are not shared between the collaborators. The UK National Security and Investment (NSI) Bill introduced in November 2020 seeks to address national security risks and includes companies working in synthetic biology and genomics[footnote 414]. It gives the UK government powers to scrutinize and intervene in business transactions and acquisitions, including investments and takeovers.
The risk from access must be balanced against the benefits of international collaboration and joint investment that is critical for research and development. In addition to the changes brought in by the NSI Bill relating to monitoring of international transactions, risk mitigation may be improved by greater pressure for reciprocity in data exchange.
Anonymisation and data protection
Global data protection legislation generally distinguishes between personal data, which benefits from protections, and anonymous data, which is subject to fewer restrictions on sharing. Individual data are protected by de-identification (removal of fields such as name, address, national insurance number) and sampling (where a representative subset of the dataset is provided, making it harder to match partial profiles to each other).[footnote 415]
The UK Data Protection Act (DPA) 2018[footnote 416] implements the EU General Data Protection Regulation (GDPR)[footnote 417] within UK law. Under GDPR, personal data is ‘any information concerning an identified or identifiable natural person’. Data that has had all direct and indirect identifiers removed, and so cannot be reidentified, is regarded as anonymised. Pseudonymised data (where direct identifiers are replaced with, for example, a reference number) can still be considered personal data under Regulation 26 of the GDPR, and its principles still apply. Pseudonymisation is effectively only a security measure and does not change the status of the data as personal data.[footnote 418]
Data protection requirements apply to all companies that process data relating to UK or EU residents regardless of whether the firm is based in the UK or EU. There are significant challenges to extra-territorial enforcement: increasing amounts of personal data are now shared internationally, and across multiple private and government bodies. Some data are processed in large internet-facing data banks, making them vulnerable to breaches. Data from different sources are increasingly combined, which may make identification easier through triangulation of information. Where breaches in anonymity take place from combining data from multiple sources, it may be difficult to determine which data led to the breach.[footnote 419]
Successful reidentification of individuals from anonymised databases has been demonstrated. The availability of data from a range of sources, such as video streaming,[footnote 420] public transport,[footnote 421] and medical treatment[footnote 422] have led to increasingly sophisticated efforts at reidentification of anonymous data. Genomic data have been previously used to identify individuals. A 2010 study found that the diagnosis codes included alongside otherwise anonymised genomic data for individuals in a GWAS study could be used to uncover individuals’ identities via electronic health records.[footnote 423]
As the volume of personal data increases, the ability to link datasets and re-identify individuals from originally anonymised data is improved.[footnote 424] The identifiability of individuals can be regarded as a function of both the features of the data and the way in which it can be accessed. Because of this, some data guardians have developed additional data access measures to ensure privacy and security.
Security
Current approaches to mitigating genomic data security risks seek to minimise access to raw data and instead offer a suite of dedicated analysis tools. The dataset for the 100,000 Genomes Project[footnote 425] is only accessible through a Trusted Research Environment (TRE) hosted by the Genomics England datacentre.[footnote 426] The TRE hosts genomic and associated clinical data, which cannot be transferred to a local device. The TRE is also preloaded with analytical tools and applications, allowing analyses to be conducted on linked, pseudonymised datasets in a controlled environment. This system of controlled access results in a higher level of data security but may add barriers to research. This controlled environment approach is increasingly being used in other databases such as the UK Secure Anonymised Information Linkage (SAIL) Databank and the Scottish Informatics Programme (SHIP).[footnote 427][footnote 428]
Whilst these repositories operate at a national level, there are efforts to facilitate global access to genetic data for research. The Global Alliance for Genomics and Health (GA4GH) is an international non-profit alliance of over 500 academic, health and commercial research organisations that aims to create frameworks and technical standards to enable the responsible, voluntary, and secure sharing of genomic and health-related data.[footnote 429] This approach could be equally applicable to non-health genomic data. The alliance attempts to respect the privacy rights of individuals while realising the public benefits of research.
Underpinning the work of GA4GH is the Framework for Responsible Sharing of Genomic and Health-Related Data.[footnote 430] This framework is founded on the principles of transparency; accountability; data quality and security; privacy, data protection and confidentiality; risk-benefit analysis; recognition and attribution; sustainability; education and training; and accessibility and dissemination.
GA4GH has created protocols, application programming interfaces (APIs), and improved file formats for sharing genomic data. Using this approach, a researcher could send queries to one or more genomic data repositories (each of which can control what information they share) or perform analyses and obtain results without accessing the raw data[footnote 431]. Such an approach could enable analysis of many datasets without needing to transfer data to one location.
Privacy-enhancing technologies are also being developed to enable easier access of data (including genomic data) whilst maintaining individual privacy.[footnote 432] Most genomic data initiatives rely on access controls to manage privacy risk.[footnote 433] Homomorphic encryption[footnote 434] could enable some processing of genomic data whilst it remains in an encrypted format,[footnote 435] providing a different privacy approach to access.[footnote 436] Whilst enhancing individual privacy, encryption schemes may reduce the inherent utility and accessibility of the data. It is therefore important to balance the burden of accessing the data with benefits to privacy.
Intellectual property
The issue of intellectual property (IP), patents and the commercialisation of genomic research touches on some of the fundamental issues with exploiting biological research. Conflicts exist between the imperatives of researchers, private companies, and the public good. Given that the major focus of genomics has historically been on health, the history of genomic intellectual property is heavily concentrated on health applications. However, there is likely to be a high amount of translation to non-health genomics in the future, with the same issues and challenges faced.
The point of patents
Patents are a form of property right that protects an invention from imitation, reflecting a bargain between society and the patent holder: they gain exclusivity over the exploitation of their invention, whilst making the principles of their invention public.[footnote 437] Patents are thought to provide benefits to innovation and competition by creating incentives for R&D, promoting the diffusion of ideas, facilitating entry to market of smaller companies, and creating markets for the trading of intellectual property. Their societal costs can include impeding subsequent innovation and creating short-term monopolies that can become entrenched.
The history of genetic patents
There has long been debate over to what extent biological materials can be patented. The US patent system originally held that “natural laws, phenomena or products of nature” could not be patented. However, the countervailing stance that biological compounds “isolated and purified” by man could be patented became accepted after an early patent case regarding epinephrine in 1900.[footnote 438]
In 1957, just a few years after the structure of DNA was identified in 1953, patents were granted on synthesized nucleoside polyphosphates and uracil; one of the four nucleotide bases used in RNA. A landmark ruling in 1980 on the patent claim regarding Pseudomonas putida (a recombinant bacterium coded to express hydrocarbon-digesting enzymes) heralded the first patent granted on a man-made living thing. The first patent for a human gene came in 1982 with CSH1, the gene for chorionic somatomammotropin. Over a thousand US patents for genes or genetic sequences then followed between 1982 and 1990.
The Human Genome Project occasioned a further controversy around genetic patenting, with initial attempts to patent the expressed sequence tags (EST) generated by the project causing disagreement between those opposed to patenting genetic material, and those seeking to capitalise on the opportunity.[footnote 439] A ruling that ESTs could only be patented when a precise biological function was identified did not exclude the possibility of patents on full-length genes, which subsequently ballooned in number, accounting for 50,000 US patents by 2009.[footnote 440]
Genetic patents after the HGP
In 1998 the European Patent Office (EPO) issued Directive 98/44/EC on the legal protection of biotechnological inventions, setting out its stance on the patentability of genetic information.[footnote 441] The directive held that “the human body, at any stage in its formation or development, including germ cells” cannot be patented. Similarly, the directive also “excludes unequivocally from patentability processes for modifying the germ line genetic identity of human beings and processes for cloning human beings”. However, the directive does not exclude plants or other biological substances from being patentable.
That same year, the US company Myriad Genetics obtained several patents on the act of isolating the sequence of two genes (BRCA1 and BRCA2), mutations of which are significant risk factors for breast, ovarian and prostate cancer. These patents gave the company the exclusive right to administer tests to detect mutations in the BRCA1 and BRCA2 gene. This was believed to reduce choice and increase costs for patients to determine if they have an increased risk of cancer. [footnote 442]
A survey conducted in the US in 2003 found that among genetic testing providers, 25 percent had stopped performing a clinical genetic test due to a patent or license, and 53 percent had decided not to develop a new clinical genetic test due to the existence of a patent or license. The overall perception of the genetic testing providers was that patents had a negative effect on the cost, access, and development of genetic tests, and on data sharing among researchers.[footnote 443]
Myriad Genetics’ patents stood until 2013, when the US Supreme Court ruled against their patents on the genetic sequence of BRCA1 and BRCA2. The Supreme Court stated that “separating that gene from its surrounding genetic material is not an act of invention” and could not be patented (though other aspects of the BRCA1/2 test remained patentable). Following the ruling, other test providers began offering the BRCA1/2 test at between $1,000 to $2,300 (versus the $4,040 charged by Myriad Genetics).[footnote 438]
The future of genetic patents
The US Supreme Court’s decision on Myriad Genetics’ patents did not settle the issue. In 2019, a draft bill was proposed in Congress that would overturn the Myriad Genetics ruling and clear the way for patenting processes and products that occur in nature.[footnote 444] The bill’s proponents argued that it would restore incentives for innovation by making the patent process more predictable. The bill’s opponents contend that the bill would herald a return to the monopolistic practices embodied by the Myriad Genetics case.
Patent issues may also affect the development of future genomic technologies. CRISPR, a genome editing technique that ‘cuts’ DNA, is governed by numerous foundational patents. Many of these are licensed by biotechnology and pharmaceutical companies, allowing them to potentially control prices for CRISPR modification.[footnote 445] The high cost of licensing these patents may disincentivise additional companies from applying the technology. High costs could also preclude CRISPR from being accessible to those on lower incomes, creating inequalities in access.
Public attitudes to genomics
The British public can see the benefits of genomic research but retain some ‘red line’ concerns regarding its use. Survey data on public attitudes to genomics suggests that the public are in general positive about some of the benefits of genomic research. However, there is some concern amongst the public. A survey by IPSOS Mori for Genomics England found that the public were concerned that genomic data might be used outside of healthcare applications in a way that could lead to a stratified society or disadvantage vulnerable people.[footnote 378] There may also be a ‘privacy paradox’[footnote 424] whereby the public feel concerned about the risk posed by genomics to data privacy, but in practice do not act to protect themselves through available safeguards.[footnote 446]
Other research indicates that the public’s willingness to donate their own DNA varies depending on who is going to use the data. Half of those surveyed were willing to donate anonymised data for use by a medical doctor, while only a quarter were willing to donate the same information to for-profit researchers.[footnote 447] Research has identified five ‘red lines’ that the public felt should be set around how genomic data (or information drawn from genomic research) should not be used. These were:
-
Genetic engineering, particularly to enhance human capability.
-
Corporate or state surveillance, particularly if it was used to penalise individuals with acute healthcare needs or single out particular groups for unfair monitoring.
-
Administrative and political usage, for example to ration access to state-funded services, or to increase controls on society.
-
Predictive insurance tests and setting personal insurance premiums.
-
Targeted marketing to increase profits, especially by large international companies.[footnote 448]
Public dialogue about the current and future uses of genomics could help to allay concerns. This can be expensive and difficult to achieve but has been successfully demonstrated previously. For example, the Ada Lovelace Institute has successfully set up ‘citizen juries’ to understand public attitudes towards different approaches to sharing of NHS data with private companies.[footnote 449] They have also worked with the Data Justice Lab to develop a guide to civic participation.
Genomic testing of newborns
Public opinion on the use of genomic sequencing for newborns is mixed. Sequencing newborns may offer earlier and more accurate identification of certain health and developmental risks than would be possible through traditional testing and monitoring. In November 2019, the Secretary of State for Health raised the possibility of sequencing the genome of newborns, with the ambition to sequence every baby born in the UK.[footnote 450] The opportunity to explore this has been reaffirmed in the latest genomic medicine implementation strategy,[footnote 85] and a dialogue on this subject has recently been published by Genomics England and the UK National Screening Committee.[footnote 76]
UK and international research suggest that public opinion is positive about the potential benefits of this, particularly for identifying rare health conditions, reducing diagnostic timescales, and improving early intervention.[footnote 451] However, participants also highlighted some concerns surrounding data safeguarding, and the future uses of genomic data. Participants also highlighted the need for accessible genetic counselling and mental health services.[footnote 76] To avoid ethical issues around unintentional findings, the Nuffield Council on Bioethics consider that “newborn screening should be limited to gene variants conferring a high risk of specific conditions that can be effectively treated or prevented in childhood”.[footnote 452]
Internationally, US and Canadian research studies have found that between 70 percent to 80 percent of parents sampled were interested in having their child’s genome sequenced.[footnote 453][footnote 454] It should be noted that the appetite for sequencing is lower than for current newborn screening technologies (94 percent) and therefore the introduction of whole genome approaches could reduce public participation in newborn screening programmes if without access to educational resources.[footnote 454]
Attitudes to genomics for forensic science
Public attitudes to the use of genomics in forensic science are mixed. People welcome the potential of genomics to facilitate more efficient and effective justice, valuing potential benefits to the detection and deterrence of crime and the prevention of miscarriages of justice[footnote 455]. However, people also raised concerns about loss of privacy, lack of consent, risks of discrimination (due to the overrepresentation of certain groups in genomic data) and risks to human rights (such as the presumption of innocence).[footnote 455]
The strength of people’s attitudes varies by level of education and exposure to law enforcement. For example, people with higher education are more likely to donate DNA samples but showed less support for local/state security agencies acting as custodians of genomic data.[footnote 456][footnote 457] They also tend to be more supportive of DNA databases of violent offenders,[footnote 458] but not those covering all citizens.[footnote 459] Groups with more exposure to criminalisation showed higher awareness of the risks, but prisoners also showed some optimism about forensic genomics as a tool to protect against wrongful accusations.[footnote 455]
Evidence to the House of Lords Science and Technology Select Committee highlighted concerns about how juries treat forensic science evidence.[footnote 460] Some experts raised the risk that DNA evidence is seen as solving everything – ‘the CSI effect’ – and that forensic genomics need to be considered as one part alongside other evidence.
People are broadly in favour of police access to genetic information. Recent polls have consistently showed that around half of the British population support a British database of all citizens’ DNA (Figure 20).[footnote 461] This is consistent with results from the US.[footnote 462] A British poll in 2019 found that 55 percent of respondents thought the police and counter-terrorism services should be able to access genomic data held by commercial companies.[footnote 463] This poll also found that 35 percent of people thought that the police could already access DNA held by these companies. These polls suggest that around half of the public are relaxed about government access to commercial DNA data, but 82 percent did not want their data to be provided to private companies.
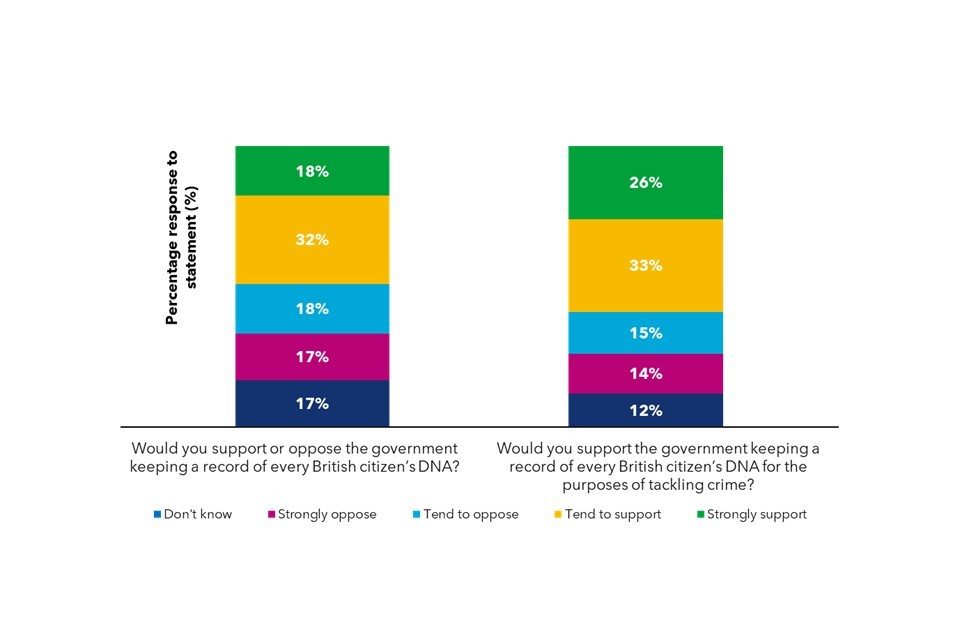
Figure 20: Public opinion on a British DNA database – generally and for use in crime, (data from YouGov, 2018).
Public engagement will continue to be important in developing the social contract around further use of genomics in forensic science. The public have views on what developments in genomics will add to the social contract in many areas, including forensic science, and should be consulted to ensure that no ‘red lines’ are crossed in how genomics is utilised for policy in the future.[footnote 378]
Summary
The general public are not always aware of the privacy risks associated with disclosing their genomic information. Policymakers could consider addressing this, perhaps taking a similar approach to campaigns that have sought to improve the public’s approach to personal information, cybersecurity and financial details. Genomic information is identifiable and is deserving of legal protection.
A clear, consistent and transparent approach to data privacy between regulators, the public and companies offering DTC genomic testing would be beneficial. Information on how the public’s genomic information can often be buried in the small print. Making it more clearly visible and understandable would enable informed consent, which is only possible if the potential consequences of using genomic data are explained.
A unified approach to regulating genomics in the UK could be considered. Genomic information is a rich resource for research that cuts across the spheres of personal data and medical information. There is a risk that developments in genomic science outpace the law. A coherent and comprehensive approach to legislating how genomic information is used could prevent this.
Current IP law may not be suitable for genomic science. A balance should be struck between rewarding innovation and benefitting the public through improved access to genomic information and treatments. The varying needs of business, academia, and public services like the NHS will need to be considered.
Clear consultation with the public and a focus on the benefits to society will need to be a priority when developing new policies in this area. The public can see the potential benefits of genomics but are also aware of its potential negative impacts on privacy, especially its use by commercial companies.
Conclusion
This report has introduced genomics for non-specialists and explored some of the current and potential future applications beyond health. Genomic science has been developing at a rapid pace. Within two decades we have progressed from the initial sequencing of the human genome to understanding the ways that thousands of genetic markers influence a range of traits. While much has been learned about the human genome and how it influences various traits, the scale of our knowledge is dwarfed by that which is not yet known. The widespread use of DNA microarray approaches has limited what we can learn from the whole genome, but the proliferation of next generation sequencing technologies is expected to provide greater insight into the workings of the human genome.
The UK has been at the forefront of genomics research for health, having developed large scale infrastructure and research capacity. Advances in genomic science and technologies are likely to be relevant to many departments across government. Given the speed at which genomic science and technology are developing, policymakers should be alert to emerging developments in genomic science, open to new opportunities for policy development and service delivery, and start considering the potential future impacts on their sectors.
Genomic science has most heavily permeated the health and medical sector. In this area it can already be used to provide early diagnosis of monogenic diseases such as Huntington’s disease and cystic fibrosis. The new NHS Genomic Medicine Service aims to build on this by integrating genomic medicine into routine clinical care. Genomic health is likely to represent the first personal exposure to genomics for many of the public. For others, they may decide to engage with direct-to-consumer genotyping companies which offer a range of genomic services.
The progress in health research and the development of genomic technology and associated infrastructure has led genomics to an inflection point where it is becoming established in fields beyond health. Genomic research is providing critical knowledge on the biological influence behind non-health traits, and is currently demonstrating the potential of its applications in non-health fields. These fields include forensic science, synthetic biology, agriculture and food, and environment and ecology, and cover a wide range of opportunities from solving criminal cold cases, to combating illegal trade, to the selective breeding of higher yield crops. Generally, the science in these areas is less advanced than in health, and the implications of applying genomic science in these fields may be more challenging.
Advances in our understanding of genomics also offers promise that this knowledge could be applied to employment, sport, education, criminal justice and insurance. While the application of genomics in these areas is in its infancy, there are a range of potential applications that may develop in the future. This may include, for example, predicting how individuals may respond to certain high-risk occupations or identifying children who may need additional support in education. Genomic prediction of non-health traits is improving, but still has some way to go to be of tangible use to professionals in these fields.
It is important to realise that some of the potential uses of genomics may not be realised in the short or even medium-term future. Many non-health traits are highly complex and genomic research in fields beyond health is in its infancy, meaning that scientific knowledge in these fields may be a decade or so behind that of health. While some genomic applications in health are both scientifically and technically possible, those in fields beyond health are generally either technically possible but reliant on a better understanding of the science, or not yet technically or scientifically possible. Nevertheless, it is increasingly important to consider how people may apply, or attempt to apply, the science and how policy and regulatory systems may need to adapt accordingly.
Numerous ethical issues surround the use of potential genomic applications in fields beyond health. Some are broad in scale and are relevant across fields, while others are specific and unique to individual fields. They cover practical issues relating to data, privacy and security, social issues like inclusion, fairness, and choice, and scientific issues such as the diversity of the populations that are represented in genomic data.
Regulation and law surrounding the use of genomics in fields beyond health are currently lacking, and in places are already behind the science. There is a risk that future scientific and technological advances could further outpace existing regulation structures, raising the possibility of genomic technologies being misused and causing harm to UK citizens. Proactive regulation that pre-empts the potential risks of future technologies would help to minimise risks and ensure that UK citizens are protected by law in terms of their genomic data privacy, anonymity, and data security. For example, the operation of direct-to-consumer genotyping companies rely on self-regulation in some areas.
It is important that government and policymakers do not just react to developments in genomics, but also take the opportunity to consider how certain aspects of genomic science may be encouraged. The UK is in a unique position to lead genomic research globally by capitalising on and further developing its existing infrastructure. It already holds a wealth of genomic data in health services and scientific studies, and has established genomic and large-scale high performance computing infrastructure to support cutting-edge research. There is an opportunity for the UK to lead internationally on regulation, law and standards relating to genomics in fields beyond health.
Key areas for policy
This report has highlighted key issues and risks that policymakers and other decision makers may wish to consider. There may also be opportunities to support the development of genomic research beyond health, and to shape its advancement and application across a broad range of sectors. These areas and opportunities can be summarised as follows:
-
Huge genomic databases containing the sequence data of millions of people are being developed to integrate genomics into healthcare. These new databases will dramatically improve the power of research studies which use them. This will also allow researchers to identify how the genome influences traits beyond health, and thereby predict an individual’s predisposition to such traits based on genomic analysis. We can already use existing databases to predict some non-health traits today, but as we accumulate vastly more comprehensive and representative genomic data, the predictive power of these tests will increase exponentially. This means that many of these predictive non-health tests will soon reach maturity, but there is very little guidance on when these tests should be used, and how the results should be interpreted.
-
A systematic assessment on the way in which genomic data could be used may be required on a sector-by-sector basis as these applications mature in the coming years. Consideration of the potential need for regulation regarding discrimination based on genomic information may also be advisable.
-
There is no overarching regulatory framework for the operation of genomic databases in the UK. Whilst genomic databases are governed by various laws, regulations and codes of practice relating to data privacy, tissue samples, and medical and research ethics, there remains an opportunity to create a unified legislative framework for their operation. This would provide clarity to both the operators of these databases, those who would volunteer their genomic data for study and the applications on the use of that data in wider settings. Such a framework would also provide a basis for transparent oversight of a hugely valuable resource. This would provide a solid foundation for the UK to build on its existing strengths in large-scale genomic databases.
-
The direct-to-consumer genomic testing market is contributing to the global growth of genomic data. But regulation of this market is ambiguous. This is partly due to the international nature of the market, and partly down to the unclear regulatory status of non-medical tests. In response, some companies have voluntarily submitted to self-regulation. However, this may not be sufficient: the data held by these companies may be vulnerable to cyberattack, their databases may be accessed by law enforcement agencies, and the test providers often do not offer supplementary guidance or interpretation of the test results to the consumer. Furthermore, there are particular risks to children and foetuses, whose rights and privacy represent complex ethical and legal dilemmas that have not yet been thoroughly considered or protected in this context. There is an opportunity to improve regulation in this area, to the benefit of both the companies and the consumer. This is particularly important as the value of genomic data increases, and the market for such testing continues to grow.
-
There is an opportunity for greater public dialogue on the current and future uses of genomics within a wider context. The national rollout of genomic medicine and the growth of direct-to-consumer testing means that more people than ever are becoming engaged with genomic science. However, there is a lack of awareness on the uses of genomic data beyond health or ancestry. Public dialogue on the value of the data beyond these examples would help to ensure that consumers are informed of the benefits and risks of seeking genomic analysis, the privacy and security implications their genomic data may create, and how the data could be reanalysed in the future, considering new scientific evidence pertaining to genomic influence on traits.
-
Genomic data is currently not fully representative of people of non-European ancestry, so genomic predictions based on this data in non-health applications risks reinforcing inequalities until this is addressed. The bias of genomic datasets towards people of European ancestry does not fully represent society, and predictions made using this data may not hold for everyone. Using inaccurate predictions to decide on important aspects of education, employment or even criminal justice risks entrenching inequality, or even contributing to it. The expanding numbers of sequences in genomic databases should go some way to addressing the issue, but it should be kept in mind when developing policy.
Acknowledgements
The Government Office for Science would like to thank the many government officials, academic and business experts and stakeholders who contributed to the work of this project, and generously provided their advice and guidance.
The project team in the Government Office for Science was led by Nancy Bailey and Dr Tom Wells and included Emily Connolly, Stephanie Croker, Melissa Jackson, Sepi Latifi, Ruth Marshall, Dr Tim Morris, Nathan Roberts, Peter Sellen, Kiran Sidhu, Talia Solel, Chris Taylor, and Simon Whitfield.
We are especially grateful to Dr Kathryn Asbury, University of York, Professor Ewan Birney, University of Cambridge and EMBL-EBI, Professor Hugh Whittall, Nuffield Council on Bioethics, and Dr Julia Wilson, Wellcome Sanger Institute, for providing expert review of content in relation to this report.
We are grateful to the following individuals for the time and input that they have provided to the report:
-
Mark Bale, Genomics England
-
Ben Allcock, Kath Bainbridge, Luke Collet-Fenson, Ross Coron, Kate Gerrand, Dafni Moschidou, Dawn O’Neill, Monika Preuss and Lauren Watson, Department of Health and Social Care
-
Gary Cook, Helen Davies, Harry Mayhew and Joseph Watts, Office for Life Sciences
-
Freddie Baker, Judith Jones, Adrian Price, Jovian Smalley, Steven Wright and Emma Young, Information Commissioner’s Office
-
Jonathan Smith, National Crime Agency
-
Callum Davies, Kirsty Faulkner, Katie Johnson, Alex MacDonald, Andy Thomson and Juliette Verdejo, Home Office
-
Lucy Barnard , William Hargreaves, Rebecca Lodge, Caron Montgomery and Sophie Rollinson, Department for Environment, Food and Rural Affairs
-
Clare Ettinghausen, Human Fertilisation and Embryology Authority
-
Thomas Corker, Adam David, James Freimanis, Natalie Hunter, Oliviero Lurcovich, StJohn Mackenzie-Boyle, Alexandra McIntyre, Eduard Mead, Jonas Nystrom, William Palmer, Jonathan Shipman, Ben West and Ben Woodham, HM Treasury
-
Catherine Joynson and Pete Mills, Nuffield Council on Bioethics
-
Gillian Atkinson, Benjamin Hepworth, Katie Hewitt and Sarah Pike, Ministry of Justice
-
Monica Aldulaimi, Ian Bradley, Mariam Orme, Deborah Sanders, Alaster Smith, Rebecca Story, Suzanne Robinson and Kelly Walker, Department for Education
-
Susan Carolin, Kathy Gammon, Mike Hill, Carl Mayers and Petra Oyston, Defence Science and Technology Laboratory
-
Andy Nisbet, Natural England
-
Mark Beaumont, Neil Davies, Stephanie von Hinke, Marcus Munafò and Lindsey Pike, University of Bristol
-
Patrick Bragoli, Rohan Kemp, Jamie Leurs, Samuel Omolade and Dehaja Senanayake, Department for Business, Energy and Industrial Strategy
-
Melike Berker, Michael Birtwistle and Nicole Huggins, Department for Digital, Culture, Media and Sport
-
Peter Burlinson, Rowan McKibbin and Jamie Parkin, Biotechnology and Biological Sciences Research Council (UKRI)
-
Prof Julia Black, London School of Economics
-
Prof Brooke Rogers, King’s College London
-
Prof Max Lu, University of Surrey
-
Prof Philip Bond, University of Manchester
-
Mike Daly and Tim Willis, Department for Work and Pensions
- Jo Harris-Roberts and Paul Willgoss, Health and Safety Executive
- Emily Smith-Woolley, University College London
Appendix
Table 4: A summary of key features of national genomic sequencing programmes for various countries.
| Australia | The Australian genomics programme, the Genomics Health Futures Mission, was launched in 2018 and is expected to last ten years. The Mission is funded (AUD $500 million; GBP £274 million) by the state-backed Medical Research Future Fund and aims to save or improve the lives of over 200,000 Australians through genomic research. Australia’s rare disease-focused approach to genomic medicine is largely facilitated by the Australian Genomics Health Alliance. This nationwide collaborative partnership reflects Australia’s federal-level system of healthcare provision, consisting of over 80 institutions across healthcare, laboratories, and academia. |
| China | By far the largest programme in terms of scale and investment in the genomic sciences is found in China. The Chinese Precision Medicine Initiative aims to sequence 100,000,000 genomes by 2030, amounting to 7.1% of the population. It is funded by a mixture of public and private investment amounting to CNY ¥59.8 billion (GBP £6.6 billion) in total for a 15-year project. Little is known internationally about the logistics of the project or who it is aiming to sequence. |
| France | The French Plan for Genomic Medicine 2025[footnote 464] (PFMG2025) was announced in 2015. The PFMG2025 has secured EUR €670 million (GBP £578 million) for the project between 2016 to 21, with approximately EUR €230 million (GBP £198 million) contributed from industry. The ten-year strategy is one of a rare disease approach, aiming to sequence approximately 235,000 genomes or genome equivalents each year to 2020. This amounts to 0.35 percent of the French population. Genomic sequencing will be performed on 20,000 rare disease patients and their parents each year, yielding a total of 60,000 genomes per year. An additional aim was that by 2020, 50,000 patients with metastatic or recurrent cancer were offered sequencing each year. This was at the whole genome, exome, or transcriptome level, contributing a further 175,000 genomes or genome equivalents. |
| Rep. of Ireland | The Republic of Ireland’s genomic medicine programme is a privately operated initiative conducted wholly by Genuity Science since 2015. It was formerly known as Genomics Medicine Ireland. In 2018 it was announced that the company had received a further EUR €350 million (GBP £302 million) in investment[footnote 465], including EUR €36 million (GBP £31 million) in state-backed financial support through the Irish Strategic Investment Fund [footnote 466]. The programme aims to collect the genomic sequences of 400,000 volunteers, amounting to around 10 percent of the population, with a focus on including patients with rare disease.[footnote 467] Volunteers to the programme receive free genomic sequencing, with between 20 to 25 percent of patients enrolled with the Genuity Rare Disease programme said to have received information of clinical significance.[footnote 468] |
| United States of America | The US genomics strategy was announced in 2015, when President Obama declared his intention to launch the first United States’ Precision Medicine Initiative, a demonstration project now known as the All of Us programme. The programme was facilitated by the National Institutes of Health, with a USD $130 million (GBP £92 million) initial fund. This has now risen to a total of USD $927 million (GBP £655 million) from a blend of public and private investment. The programme seeks to collect genomic data from approximately 1,000,000 participants by 2022. Enrolment into the programme began in 2018, with the first genomic data was obtained in 2019. The study aims to capture genomic data that is reflective of the general national population demographic, not necessarily targeting those with rare diseases or cancer as seen with other initiatives. It also aims to gather a range of holistic data from participants in addition to their genomic data. This includes voluntary submission of participant electronic health records, physiological measurements such as height and weight, and third-party fitness data obtained via Fitbit.[footnote 469] |
Glossary
Acronyms
| Term | Definition | |
|---|---|---|
| ABI | Association of British Insurers | |
| ACE | Angiotensin I-Converting Enzyme | |
| AFB | Age at First Birth | |
| AI | Artificial Intelligence | |
| ASB | Anti-Social Behaviour | |
| AUD | Australian Dollar | |
| AUDIT | Alcohol Use Disorder Identification Test | |
| BMI | Body Mass Index | |
| CLOUD | Clarifying Lawful Overseas Use of Data | |
| CNY | Chinese Yuan | |
| COPO | Crime (Overseas Production Orders) | |
| COVID | Corona Virus Disease | |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats | |
| CSI | Crime Scene Investigation | |
| DHSC | Department for Health and Social Care | |
| DIY | Do it Yourself | |
| DTC | Direct-to-Consumer | |
| EBI | European Bioinformatics Institute | |
| EC | European Communities | |
| ECG | Electrocardiogram | |
| EEA | European Economic Area | |
| EPO | Erythropoietin | |
| EST | Expressed Sequence Tags | |
| EU | European Union | |
| EUR | Euro | |
| FBI | Federal Bureau of Investigation | |
| FCA | Financial Conduct Authority | |
| FDA | Food and Drug Administration | |
| FIMS | International Federation of Sports Medicine | |
| GBP | British Pound Sterling | |
| GDPR | General Data Protection Regulations | |
| GEBV | Genomic Estimated Breeding Value | |
| GINA | Genetic Information Non-Discrimination Act | |
| GM | Genetically Modified | |
| GMO | Genetically Modified Organism | |
| GMS | Genomic Medicine Service | |
| GP | General Practitioner | |
| GWAS | Genome-Wide Association Study | |
| HFEA | Human Fertilisation and Embryology Authority | |
| HGC | Human Genetics Commission | |
| HGP | Human Genome Project | |
| HIV | Human Immunodeficiency Virus | |
| HSE | Health and Safety Executive | |
| HTT | Huntingtin gene | |
| ICO | Information Commissioner’s Office | |
| IP | Intellectual Property | |
| IPSOS | Institut de Publique Sondage d’Opinion Secteur | |
| IQ | Intelligence Quotient | |
| IVD | In vitro Diagnostic | |
| JCVI | J. Craig Venter Institute | |
| JPEG | Joint Photographic Experts Group | |
| MHRA | Medicines and Healthcare products Regulatory Agency | |
| MRI | Magnetic Resonance Imaging | |
| mRNA | Messenger RNA | |
| NDNAD | National DNA Database | |
| NGS | Next Generation Sequencing | |
| NHGRI | National Human Genome Research Institute | |
| NHS | National Health Service | |
| NIHR | National Institute for Health Research | |
| PCR | Polymerase Chain Reaction | |
| Portable Document Format | ||
| PPAR | Peroxisome Proliferator-Activated Receptor | |
| PRS | Polygenic Risk Score | |
| PS | Polygenic Score | |
| RNA | Ribonucleic Acid | |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 | |
| SNP | Single Nucleotide Polymorphism | |
| STR | Short Tandem Repeat | |
| SUD | Substance Use Disorder | |
| TYR | Tyrosinase | |
| UK | United Kingdom | |
| UKRI | UK Research and Innovation | |
| US | United States | |
| USA | United States of America | |
| USD | United States Dollar | |
| WADA | World Anti-Doping Agency | |
| WES | Whole Exome Sequencing | |
| WGS | Whole Genome Sequencing |
Terminologies
| Term | Definition | |
|---|---|---|
| Adenine | See ‘nucleotide’. | |
| Allele | An alternative form of a gene, created by mutation. | |
| Amino acid | The organic compounds which make up the majority of proteins. | |
| Autosome | The 22 pairs of human chromosomes that are not sex-linked. | |
| Base | See ‘nucleotide’. | |
| Biobank | A biorepository that stores biological material for research, such as DNA or tissue samples. | |
| Chromatin | A compacted form of DNA and protein which forms the essential chromosome. | |
| Chromosome | Strands of X-shaped compacted DNA that are found in almost all cells. Humans have 46 chromosomes, subdivided into 23 pairs. | |
| Coding DNA | A DNA sequence which codes for amino acids. See ‘amino acid’, ‘gene’ and ‘exome’. | |
| Cytosine | See ‘nucleotide’. | |
| Diploid | A cell or organism that possess two copies of each chromosome. Nearly all human cells are diploid, and thereby contain 23 pairs of chromosomes. | |
| Direct-to-Consumer | Private companies which offer genomic analysis directly to the consumer, for ancestry, health, or non-health purposes. | |
| Dominant | An allele of a gene which expresses itself more strongly than other alleles of the same gene. | |
| Epigenetic | Refers to changes in gene activity which are not caused by changes to the DNA sequence, but instead how the DNA is packaged/spatially arranged. See ‘histones’ and ‘chromatin’. | |
| Epistasis | The interaction between two or more different genes the same phenotype. | |
| Exome | Collective term for the protein-coding sections of the genome. | |
| Fingerprinting | The comparative use of DNA samples to identify samples and suspects, based off similarities in DNA STR locations. See ‘STR’. | |
| Gamete | See ‘germ cell’. | |
| Gene | A section of protein-coding DNA which contains the sequence pertaining to an entire protein. | |
| Genome | An entire haploid set of chromosomes of an organism. In humans, this consists of 23 chromosomes. | |
| Genome/ Gene Editing | The process of inserting, deleting, modifying, or replacing DNA within a living organism. Gene editing refers to this process within the context of a single gene, genome editing within the larger context of the organism. | |
| Genome-wide Association Study | A large-scale genomic study which aims to associate specific genetic variations with particular traits, such as disease. | |
| Genomic Estimated Breeding Value | A score used within agriculture and livestock farming that quantifies the genetic contribution towards a specific trait. These can be used to improve selective breeding processes (see ‘genomic selection’). GEBVs are largely analogous to ‘polygenic scores’. | |
| Genomic Selection | The optimised process of selecting agricultural/livestock breed stock based on genomic information. Often utilises ‘GEBV’s. | |
| Genotype | An individual’s collection of genes. Can refer to both their whole genome, and specific genes/alleles. | |
| Germ cell | An immature biological cell that later develops into the gametes of an organism. Human germ cells mature into the human gametes: sperm and eggs. | |
| Germline mutation | Specifically refers to a genetic mutation occurring only within an organism’s ‘germ cells’. Germline mutations can pass between generations. | |
| Guanine | See ‘nucleotide’. | |
| Haploid | A cell or organism that possesses a single copy of each chromosome. Human ‘germ cells’ are haploid and contain 23 chromosomes. | |
| Heritability | A population-level statistic that estimates the degree of variation in a trait that is due to genetic variation between individuals in that population. | |
| Histones | The proteins responsible for the packaging of chromatin. Reversible modification of histones can effectively switch genes on or off. See ‘chromatin’ and ‘epigenetics’. | |
| Imputation | A process of predicting genomic variants by the presence of others, as determined through DNA microarray. | |
| Messenger RNA | An intermediate transcribed copy of DNA, which undergoes the translation process within the protein synthesis pathway. See ‘transcription’ and ‘translation’ for functional terms. | |
| Microarray | A technique to survey a DNA sequence for thousands of known gene variants quickly and cheaply. See ‘imputation’. | |
| Monogenic | A trait which is caused by a single gene. | |
| Mutation | An unintentional alternation of the standard nucleotide sequence. Can occur naturally over time (aging) or as a consequence of environment (such as cigarette smoke). | |
| Next Generation Sequencing | A sequencing technique which became available in the mid-2000’s. This is a high-throughput, scalable, and quick method compared to Sanger sequencing. | |
| Non-coding DNA | A DNA sequence which does not code for amino acids. | |
| Nucleosomes | Refers to a unit of DNA which is wrapped around a histone. | |
| Nucleotide | The basic building block of nucleic acids, such as DNA and mRNA. The four nucleotides of DNA are adenine, thymine, guanine, and cytosine. They may also be known as bases. | |
| Penetrant | In genomics, it refers to the proportion of individuals which possess a gene variant and then subsequently express the associated trait. | |
| Phenotype | The observable physical properties of an organism, occurring through interaction of the ‘genotype’ with the environment. An example is an individual’s appearance. | |
| Pleiotropy | The concept that one gene can lead to multiple phenotypes or manifestations. Some types of albinism arise through pleiotropy of the melanin gene. | |
| Polygenic | A trait which is caused by a several genes, ranging from two to over thousands. | |
| Polygenic Score | A calculated number that summarises the estimated effect of many genetic variants on an individual’s phenotype. Commonly used within the context of disease, such as breast cancer risk. | |
| Polymerase Chain Reaction | A laboratory technique used to make copies of a specific DNA strand. | |
| Protein | A large biomolecule which is comprised of amino acids. Cells and tissues are largely built with proteins, and they are the second most abundant compound in the body, following water. | |
| Recessive | An allele of a gene which expresses itself weakly compared to other alleles of the same gene. Traits may still be caused by these mutated alleles if the individual exclusively possesses mutated copies of the allele. | |
| Ribonucleic Acid | See ‘messenger RNA’. | |
| Sanger Sequencing | One of the first methods of sequencing DNA. It was used in the Human Genome Project, which concluded in the early 2000’s. | |
| Single Nucleotide Polymorphism | A DNA sequence variation occurring when a single nucleotide changes. Caused by mutation, new mutated versions of the same gene are known as alleles. | |
| Somatic mutation | A mutation in a DNA sequence that occurs after conception and in cells not related to sexual function (gametes). Mutations are not passed to descendants. | |
| Thymine | See ‘nucleotide’. | |
| Transcription | Part of the initial process by which DNA contains the instructions to make proteins. Transcription refers to the process of making an mRNA copy of a section of DNA. See ‘translation’. | |
| Translation | Part of the latter process by which DNA contains the instructions to make proteins. Translation refers to the processing of mRNA into an amino acid sequence, forming a protein. See ‘transcription’. | |
| Twin Studies | A type of study which examines the development of traits/phenotypes between identical and non-identical twins, thereby determining how much of a trait is genomically influenced, and how much is environmentally influenced. |
-
National Human Genome Research Institute (2020) DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Accessed 17 June 2021. ↩
-
Young, A (2019) Solving the missing heritability problem PLoS Genetics, 15(6), DOI: 10.1371/journal.pgen.1008222 ↩ ↩2
-
Yengo, L (2018) Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry Human Molecular Genetics, 27(20), DOI: 10.1093/hmg/ddy271 ↩ ↩2
-
Mills, M (2019) A scientometric review of genome-wide association studies Communications Biology, Communications Biology, 2(9), DOI: 10.1038/s42003-018-0261-x ↩ ↩2 ↩3 ↩4
-
Morris, T (2020) Can education be personalised using pupils’ genetic data? eLife, 9, DOI: 10.7554/eLife.49962 ↩ ↩2
-
DHSC, BEIS, OLS, and Lord Bethell of Romford (2020) Genome UK: the future of healthcare.. Accessed 17 June 2021. ↩
-
Bergin, J (2020) Direct-to-Consumer Genetic Testing: Global Markets and Technologies. BCC Research. Accessed 18 June 2021. ↩ ↩2
-
Home Office (2021) National DNA Database statistics Accessed 18 June 2021. ↩ ↩2
-
Resnick, B (2018) How your third cousin’s ancestry DNA test could jeopardize your privacy. Vox. Accessed 8 October 2021. ↩
-
International Society of Genetic Genealogy Wiki (2021) Autosomal DNA statistics. Accessed 8 October 2021. ↩
-
Jang, K (1996) Heritability of the big five personality dimensions and their facets: a twin study. Journal of personality, 64(3), 577-592, DOI: 10.1111/j.1467-6494.1996.tb00522.x ↩ ↩2
-
Suter, S. M. (2018). GINA at 0 years: the battle over ‘genetic information’ continues in court. Journal of Law and the Biosciences, 495-526, 5(3), DOI: https://doi.org/10.1093/jlb/lsz002 ↩
-
Enriquez, J (2012) Genetically enhanced Olympics are coming. Nature, 487(297), DOI: 10.1038/487297a ↩ ↩2
-
Tanisawa, K (2019) Sport and exercise genomics: The FIMS 2019 consensus statement update British Journal of Sports Medicine, 969-975, 54(16), DOI: 10.1136/bjsports-2019-101532 ↩ ↩2 ↩3
-
Regalado, A (20 8) ‘DNA tests for IQ are coming, but it might not be smart to take one. MIT Technology review. Accessed 25 June 2021. ↩ ↩2
-
Agrawal, A (2012) The genetics of addiction—a translational perspective Translational psychiatry, 2(7), DOI:10.1038/tp.2012.54 ↩ ↩2
-
Howard, R (2016) Genetic Testing Model for CI: If Underwriters of Individual Critical Illness Insurance Had No Access to Known Results of Genetic Tests. Canadian Institute of Actuaries. Accessed 21 June 2021. ↩ ↩2
-
House of Commons Science and Technology Committee (2021) Direct-to-consumer genomic testing – first report. Accessed 28 June 2021. ↩ ↩2 ↩3 ↩4 ↩5
-
The concept of “junk DNA” arose from a lack of knowledge about non-coding DNA and what it does. Non-coding DNA used to be thought of as old DNA which did not work or do much anymore, but the term is gradually disappearing from scientific literature as we improve our understanding of the regulatory functions of non-coding DNA within the context of the wider genome. ↩
-
Autosomal refers to the fact that the mutation occurs on a non-sex specific chromosome, also known as the autosome. Mutations on X or Y chromosomes (which determine sex) display slightly different inheritance patterns depending on the sex of the child inheriting the gene. More information on sex-specific traits can be found here ↩
-
Marouli, E (2017) Rare and low-frequency coding variants alter human adult height. Nature, 542(7640), DOI: 10.1038/nature21039 ↩
-
Lee, J (2018) Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics, 50(8), DOI: https://doi.org/10.1038/s41588-018-0147-3 ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8 ↩9 ↩10 ↩11
-
Naumova O (2019) Effects of early social deprivation on epigenetic statuses and adaptive behavior of young children: A study based on a cohort of institutionalized infants and toddlers PLoS ONE, 14(3), DOI: 10.1371/journal.pone.0214285 ↩
-
Gottschalk, M (2020) Epigenetics Underlying Susceptibility and Resilience Relating to Daily Life Stress, Work Stress, and Socioeconomic Status. Gottschalk, M (2020) Epigenetics Underlying Susceptibility and Resilience Relating to Daily Life Stress, Work Stress, and Socioeconomic Status. Front. Psychiatry, 11(163), DOI: 10.3389/fpsyt.2020.00163 Front. Psychiatry, 11(163), DOI: 10.3389/fpsyt.2020.00163 ↩
-
McCartney, D (2018) Epigenetic signatures of starting and stopping smoking. EBioMedicine, 37, DOI: 10.1016/j.ebiom.2018.10.051 ↩
-
National Human Genome Research Institute (2018) What is the Human Genome Research Project? Accessed 12 May 2021. ↩
-
Collins, F (2003) The Human Genome Project: Lessons from Large-Scale Biology Science, 300(5617), DOI: 10.1126/science.1084564 ↩
-
National Human Genome Research Institute (2012) International HapMap project Accessed 21 May 2021. ↩
-
National Human Genome Research Institute (2020) The Encyclopedia of DNA Elements (ENCODE). Accessed 17 June 2021. ↩
-
Centre of Excellence for Engineering Biology (2021) About the Centre of Excellence for Engineering Biology. Accessed 17 June 2021. ↩
-
The Darwin Tree of Life Project (2021) Homepage. Accessed 5th October 2021. ↩ ↩2
-
‘Adult’ in this context spans the time from birth to death. Umbilical cord blood obtained during birth would fall under an ‘adult’ definition. ↩
-
Sanger, F (1977) DNA sequencing with chain-terminating inhibitors. Proceedings of the Natural Academy of Sciences of the USA, 74(12), DOI: 10.1073/pnas.74.12.5463 ↩
-
Heather, J (2016) The sequence of sequencers: The history of sequencing DNA Genomics, 107(1), DOI: 10.1016/j.ygeno.2015.11.003 ↩
-
University of Cambridge (2019) Collaboration and Impact: Solexa Sequencing Accessed 30 July 2021. ↩
-
Voelkerding, K (2009) Next-Generation Sequencing: From Basic Research to Diagnostics Clinical Chemistry, 55(4), DOI: 10.1373/clinchem.2008.112789 ↩
-
Oxford Nanopore (2021) Company History Accessed 30 July 2021. ↩
-
Mina-Vargas, A (2017) Heritability and GWAS Analyses of Acne in Australian Adolescent Twins Twin Research and Human Genetics, 20(6), DOI: 10.1017/thg.2017.58 ↩
-
Colvert, E (2015) Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample JAMA Psychiatry, 72(5), DOI: 10.1001/jamapsychiatry.2014.3028 ↩
-
Elks, C (2012) Variability in the heritability of body mass index: a systematic review and meta-regression Front. Endocrinol, DOI: 10.3389/fendo.2012.00029 ↩
-
Kendler, K (2006) A Swedish National Twin Study of Lifetime Major Depression Am J Psychiatry, 163(1), DOI: 10.1176/appi.ajp.163.1.109 ↩
-
Lin, BD (2015) Heritability and Genome-Wide Association Studies for Hair Color in a Dutch Twin Family Based Sample Genes (Basel), 6(3), DOI: 10.3390/genes6030559 ↩
-
Medland, S (2009) Genetic influences on handedness: data from 25,732 Australian and Dutch twin families. Neuropsychologia, 47(2), DOI: 10.1016/j.neuropsychologia.2008.09.005 ↩
-
Plomin, R (2015) Genetics and intelligence differences: five special findings Mol Psychiatry, 20(1), DOI: 10.1038/mp.2014.105 ↩ ↩2
-
Haworth, C (2010) The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry, 15, DOI: 10.1038/mp.2009.55 ↩ ↩2 ↩3
-
Gingras, B (2015) Defining the biological bases of individual differences in musicality Phil. Trans. R. Soc. B, 370, DOI: 10.1098/rstb.2014.0092 ↩
-
Sullivan, P (2003) Schizophrenia as a Complex Trait: Evidence from a Meta-analysis of Twin Studies. Arch Gen Psychiatry, 60(12), DOI: 10.1001/archpsyc.60.12.1187 ↩
-
Plomin, R (1977) Genotype–environment interaction and correlation in the analysis of human behavior. Psychological Bulletin, 84(2): 309–322. DOI: 10.1037/0033-2909.84.2.309. ↩
-
Visscher, P (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet, 101(1), DOI: 10.1016/j.ajhg.2017.06.005 ↩
-
EBI GWAS Catalogue data available here and here Accessed 17 June 2021. ↩
-
Silventoinen, K (2012) Heritability of Adult Body Height: A Comparative Study of Twin Cohorts in Eight Countries Twin Research and Human Genetics, 6(5), DOI: 10.1375/twin.6.5.399 ↩
-
Yang, J (2010) Common SNPs explain a large proportion of the heritability for human height Nature Genetics, 42, DOI: 10.1038/ng.608 ↩
-
Duncan, L (2019) Analysis of polygenic risk score usage and performance in diverse human populations Nature Communications, 3328, DOI: 10.1038/s41467-019-11112-0 ↩
-
Martin, A (2017) Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations Am J Hum Genet, 10(4), DOI: 10.1016/j.ajhg.2017.03.004 ↩
-
Martschenko, D (2019) Genetics and Education: Recent Developments in the Context of an Ugly History and an Uncertain Future AERA Open, 5(1), DOI: 10.1177/2332858418810516 ↩ ↩2
-
Tam, V (2019) Benefits and limitations of genome-wide association studies Nat. Rev. Genet, 20, DOI: 0.1038/s41576-019-0127-1 ↩
-
PREPRINT: Wainschtein, P (2021) Recovery of trait heritability from whole genome sequence data bioRxiv, DOI: 10.1101/588020 ↩
-
The UK10K Consortium (2015) The UK10K project identifies rare variants in health and disease Nature, 526, DOI: 10.1038/nature14962 ↩
-
Maas, P (2016) Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States JAMA oncology, 2(10), DOI: 10.1001/jamaoncol.2016.1025 ↩
-
Mosley, J (2020) Predictive Accuracy of a Polygenic Risk Score Compared with a Clinical Risk Score for Incident Coronary Heart Disease JAMA, 323(7), DOI: 10.1001/jama.2019.21782 ↩
-
Hellwege, J (2018) Population Stratification in Genetic Association Studies . Curr Protoc Hum Genet, 95, DOI: 10.1002/cphg.48 ↩
-
Price, A (2010) New approaches to population stratification in genome-wide association studies Nat Rev Genet, 11, DOI: 10.1038/nrg2813 ↩
-
Lawson, D (2019) Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity? Human Genetics, 139, DOI: 10.1007/s00439-019-02014-8. ↩
-
Kong, A (2018) The nature of nurture: Effects of parental genotypes Science, 359(6374) DOI: 10.1126/science.aan6877 ↩
-
Morris, T (2020) Population phenomena inflate genetic associations of complex social traits. Science Advances, 6(16), DOI: 10.1126/sciadv.aay0328 ↩ ↩2
-
HM Government (2019) Advancing our health: prevention in the 2020s – consultation document Accessed 26 July 2021 ↩
-
Bell, J and Office for Life Sciences (2017) Life Sciences Industrial Strategy Accessed 17 June 2021. ↩
-
Genomics England (2013) The 100,000 Genomes Project Accessed 17 June 2021. ↩
-
Genomics England (2013) About Genomics England Accessed 17 June 2021. ↩
-
Genomics England (2018) The UK has sequenced 100,000 whole genomes in the NHS Accessed 17 June 2021. ↩
-
Davies, C, Chief Medical Officer (2016) Annual Report of the Chief Medical Officer 2016: Generation Genome Accessed 17 June 2021. ↩
-
UKRI (2020) UK Biobank. Accessed 21 June 2021. ↩
-
UKRI (2021) Accelerating detection of disease challenge. Accessed 17 June 2021. ↩
-
NIHR Bioresource (2021) What we do. Accessed 17 June 2021. ↩
-
Department of Health and Social Care (2021) The UK Rare Diseases Framework. Accessed 17 June 2021. ↩
-
UK National Screening Committee (2021) Implications of whole genome sequencing for newborn screening (executive summary). Accessed 28 July 2021. ↩ ↩2 ↩3
-
Nussinov, R (2021) A new precision medicine initiative at the dawn of exascale computing. Signal Transduction and Targeted Therapy, 6(1) DOI: 10.1038/s41392-020-00420-3 ↩
-
Kovanda, A (2021) How to design a national genomic project—a systematic review of active projects. Human Genomics, 15(1) DOI: 10.1186/s40246-021-00315-6 ↩ ↩2
-
D’Cruz, R (2020) Laboratory Testing Methods for Novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2 Frontiers in Cell and Developmental Biology, 8, DOI: 10.3389/fcell.2020.00468 ↩
-
Kames, J (2020) Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design. Scientific Reports, 10(1), DOI: 10.1038/s41598-020-72533-2 ↩
-
Cono, J (2021) mRNA COVID-19 Vaccines: An Incredible Feat of Genomic Technology. Centers for Disease Control and Prevention. Accessed 18 June 2021. ↩
-
Challen, R (2021) Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ, 372(n579), DOI: 10.1136/bmj.n579 ↩
-
Ott, A (2020) Monitoring wastewater for COVID-19.Parliamentary Office for Science and Technology. Accessed 18 June 2021. ↩
-
Korber, B (2020) Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell, 182(4), DOI: 10.1016/j.cell.2020.06.043 ↩
-
DHSC, BEIS and OLS (2021) Genome UK: 2021 to 2022 implementation plan. Accessed 18 June 2021. ↩ ↩2
-
Gardy, J (2018) Towards a genomics-informed, real-time, global pathogen surveillance system. Nature Reviews Genetics, 19(1), DOI: 0.1038/nrg.2017.88 ↩
-
Animal and Plant Health Agency (2020) APHA Annual Science and Evidence Review 2019. Accessed 21 June 2021. ↩
-
Radhakrishnan, G (2019) MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biology, 17(1), DOI: 10.1186/s12915-019-0684-y ↩
-
Brown, E (2019) Use of Whole-Genome Sequencing for Food Safety and Public Health in the United States. Foodborne Pathogens and Disease, 16(7), DOI: 10.1089/fpd.2019.2662 ↩
-
Plewniak, F (2018) A Genomic Outlook on Bioremediation: The Case of Arsenic Removal. Frontiers in Microbiology, 9(820), DOI: 0.3389/fmicb.2018.00820 ↩
-
Zahedi, A (2021) Wastewater-based epidemiology—surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitology Research, DOI: 10.1007/s00436-020-07023-5 ↩
-
Bibby, K (2019) Metagenomics and the development of viral water quality tools. npj Clean Water, (2019), 2(1), DOI: 10.1038/s41545-019-0032-3 ↩
-
Behzad, H (2015). Challenges and opportunities of airborne metagenomics. Genome biology and evolution, 7(5), DOI: 10.1093/gbe/evv064 ↩
-
Cao C (2014) Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ Sci Technol, 48, DOI: 10.1021/es4048472 ↩
-
Alexander, J (2017) Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol, 14(6), DOI: 10.1038/nrgastro.2017.20 ↩
-
Dominguez-Bello , M (2016) Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nature Medicine, 22, DOI: 10.1038/nm.4039 ↩
-
Pennisi, E (2019) Evidence mounts that gut bacteria can influence mood, prevent depression. Science, accessed 30 July 2021. ↩
-
Sharma, A (2020) The Future of Microbiome-Based Therapeutics in Clinical Applications. Clinical Pharmacology & Therapeutics, 107(1), DOI: 10.1002/cpt.1677 ↩
-
Almeida, A (2019) A new genomic blueprint of the human gut microbiota. Nature, 568, DOI: 10.1038/s41586-019-0965-1 ↩
-
EMBL-EBI (2021) MGnify. Accessed 30 July 2021. See also Mitchell, A (2019) MGnify: the microbiome analysis resource in 2020. Nucleic Acids Research, 48(D1), DOI: 10.1093/nar/gkz1035 ↩
-
Forster, S (2019) A human gut bacterial genome and culture collection for improved metagenomic analyses. Nature Biotechnology, 37, DOI: 10.1038/s41587-018-0009-7 ↩
-
A correlation has been identified between the two. See Kim, S (2020) Altered Gut Microbiome Profile in Patients With Pulmonary Arterial Hypertension. Hypertension, 75, DOI: 10.1161/HYPERTENSIONAHA.119.14294 ↩
-
Muscogiuri, G (2019) Gut microbiota: a new path to treat obesity. International Journal of Obesity Supplements, 9, DOI: 10.1038/s41367-019-0011-7 ↩
-
Bastiaanssen, T (2019) Making Sense of … the Microbiome in Psychiatry. Int J Neuropsychopharmacol, 22(1), DOI: 10.1093/ijnp/pyy067 ↩
-
Friend, L (2018) Direct-to-consumer genetic testing: Opportunities and risks in a rapidly evolving market. KPMG. Accessed 18 June 2021. ↩
-
Regalado, A (2019) More than 26 million people have taken an at-home ancestry test. MIT Technology Review. Accessed 18 June 2021. ↩ ↩2
-
Yudell, M (2016) Taking race out of human genetics. Science, 351(6273), DOI: 10.1126/science.aac4951 ↩
-
Jelenkovic, A (2016) Genetic and environmental influences on height from infancy to early adulthood: An individual-based pooled analysis of 45 twin cohorts. Scientific Reports, 6, DOI: 10.1038/srep28496 ↩
-
Weedon, MN (2021) Use of SNP chips to detect rare pathogenic variants: retrospective, population based diagnostic evaluation. BMJ, 372(214), DOI: 10.1136/bmj.n214 ↩
-
Veritas Genetics (2020) Whole genome sequencing and interpretation. Accessed 18 June 2021. ↩
-
House of Commons Science and Technology Committee (2020) Oral evidence: Commercial Genomics, HC 140. Accessed 18 June 2021. ↩ ↩2
-
23andMe (2020) Written Evidence Submitted by 23andMe to the Science and Technology Committee’s Commercial Genomics Inquiry (COG0002). Accessed 18 June 2021. ↩
-
Karavani, E (2019) Screening Human Embryos for Polygenic Traits Has Limited Utility. Cell, 179(6), DOI: 10.1016/j.cell.2019.10.033. ↩
-
Turley, P (2021). Problems with Using Polygenic Scores to Select Embryos. New England Journal of Medicine, 385(1), DOI: 10.1056/NEJMsr2105065. ↩
-
Phillips, A (2016) Only a click away — DTC genetics for ancestry, health, love…and more: A view of the business and regulatory landscape. Applied & Translational Genomics, 8, DOI: 10.1016/j.atg.2016.01.001 ↩
-
Genomelink. [Upload DNA data and know more about yourself.] (https://genomelink.io/) Accessed 18 June 2021. ↩
-
Genomelink. [The 17 Best DNA Upload Sites for Additional Analysis on Raw DNA Data File in 2020.] (https://blog.genomelink.io/posts/best-raw-dna-data-upload-sites) Accessed 18 June 2021. ↩
-
Moray, N (2017). Paternity testing under the cloak of recreational genetics.European Journal of Human Genetics, 25(6), DOI: 10.1038/ejhg.2017.31 ↩
-
In England, Wales and NI, whether or not the right of a child or parent to consent to a test is determined by the “Gillick test” whereby “the parental right to determine whether or not their minor child below the age of sixteen will have medical treatment terminates if and when the child achieves sufficient understanding and intelligence to understand fully what is proposed” – see discussion in Griffith, R (2016) What is Gillick competence?. Human vaccines & immunotherapeutics, 12(1), DOI: 10.1080/21645515.2015.1091548. See also guidance on the Human Tissue Act (HTA) regarding genetic testing. ↩
-
Nuffield Council on Bioethics (2010) Medical profiling and online medicine: the ethics of ‘personalised healthcare’ in a consumer age.Accessed 28 July 2021. ↩
-
Human Fertilisation & Embryology Authority (2020) DNA tests are the Christmas gift that keeps on giving, but unexpected results can bring you more than you bargained for. Accessed 18 June 2021. ↩
-
The Wall Street Journal (2019) [The Patriot Act Goes Too Far.] (https://www.wsj.com/articles/the-patriot-act-goes-too-far-11572209177)Accessed 18 June 2021. ↩
-
U.S. Department of Justice (2004) Report from the field: the USA PATRIOT Act at work. Accessed 18 June 2021. ↩
-
Dove, E (2015) Genomic cloud computing: legal and ethical points to consider. European Journal of Human Genetics, 23(10), DOI: 10.1038/ejhg.2014.196 ↩
-
US Department of Justice (2019) US and UK Sign Landmark Cross-Border Data Access Agreement to Combat Criminals and Terrorists online.Accessed 18 June 2021. ↩
-
Wellcome Sanger Institute (2019) Commercial genomics inquiry response. Accessed 18 June 2021. ↩
-
See e.g., Ofcom’s 2020/21 report on Adults’ media use and attitudes. Accessed 18 June 2021. ↩
-
Financial Times (2017) Biotechnology: the US-China dispute over genetic data. Accessed 18 June 2021. ↩
-
Gryphon Scientific and Rhodium Group (2019) China’s Biotechnology Development: The Role of US and other Foreign Engagement. Accessed 18 June 2021. ↩
-
European Commission (2019) What rules apply if my organisation transfers data outside the EU? Accessed 18 June 2021. ↩
-
Lynch, D (2017) Biotechnology: the US-China dispute over genetic data. The Financial Times. Accessed 28 July 2021. ↩
-
Selita, F. (2019). Genetic data misuse: risk to fundamental human rights in developed economies. Legal Issues Journal, 7(1). ↩
-
Defined as providing a medical diagnosis or predisposition to a medical condition. ↩
-
The Medical Devices (Amendment etc.) (EU Exit) Regulations 2019 ↩
-
The Secretary of State for Health or other appropriate authority can amend these regulations according to the Medicines and Medical Devices Act (2021). ↩
-
Kirkpatrick, B (2017) Ancestry Testing and the Practice of Genetic Counseling. J Genet Couns, 26(1), DOI: 10.1007/s10897-016-0014-2 ↩
-
U.S. Food and Drug Administration (FDA). Direct-to-Consumer Tests. Accessed: December 2019. ↩ ↩2
-
See for example Tiller, J (2018). Regulation of Internet-based genetic testing: challenges for Australia and other jurisdictions. Frontiers in Public Health, 6(24), DOI: 10.3389/fpubh.2018.00024 ↩
-
See Consumer Protection Act 1987 and the Consumer Rights Act 2015 for non-diagnostic test quality and safety standards, UK General Data Protection Act 2018 for data use, [Human Tissue Act 2004] (https://www.legislation.gov.uk/ukpga/2004/30/contents) for sample processing and consent, and the Consumer Protection from Unfair Trading Regulations 2008 and the Business Protection from Misleading Marketing Regulations 2008 for advertising standards. All pages accessed 28 July 2021. ↩
-
Human Genetics Commission (2010) A Common Framework of Principles for direct-to-consumer genetic testing services. Accessed 18 June 2021. ↩
-
Hall (2017) Transparency of genetic testing services for ‘health, wellness and lifestyle’: analysis of online prepurchase information for UK consumers. European Journal of Human Genetics, 25, DOI: 10.1038/ejhg.2017.75 ↩
-
Jonsson, H (2021). Differences between germline genomes of monozygotic twins. Nat Genet, 53, DOI: 10.1038/s41588-020-00755-1. ↩
-
Fingerprints and iris patterns, which are used for biometric identification, are, like DNA, unique to the individual (even identical twins have different fingerprints) but unlike DNA they cannot be used to infer relatedness. ↩
-
In the UK, DNA evidence can be provided to prove a family relationship in immigration rules, but it is voluntary to do so. ↩
-
Crown Prosecution Service (2017) Legal guidance on DNA-17 Profiling. Accessed 18 June 2021. ↩
-
[Crime and Security Act 2010] (https://www.legislation.gov.uk/ukpga/2010/17/enacted) ↩
-
See for example: BBC (2018) How familial DNA trapped a murderer for the first time. Accessed 18 June 2021. ↩
-
Tillmar, A (2021) Getting the conclusive lead with investigative genetic genealogy – A successful case study of a 16 year old double murder in Sweden. FSI Genetics, 53, DOI: 10.1016/j.fsigen.2021.102525 ↩
-
Verogen (2021) Preparation Kits. Accessed 30 July 2021. ↩
-
Mushailov V (2015) Assay development and validation of an 8-SNP multiplex test to predict eye and skin coloration. J Forensic Sci, 60(4), DOI: 10.1111/1556-4029.12758 ↩
-
Walsh, S (2017) [Global skin colour prediction from DNA.] (https://link.springer.com/article/10.1007/s00439-017-1808-5) Human Genetics, 136, DOI: 10.1007/s00439-017-1808-5 ↩
-
For prediction of height see: Liu, F (2019) Update on the predictability of tall stature from DNA markers in Europeans. Forensic Science International: Genetics, 42, DOI: 10.1016/j.fsigen.2019.05.006 ↩
-
Aliferi, A (2018) DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Science International: Genetics, 37, DOI: 10.1016/j.fsigen.2018.09.003. ↩
-
Van Noorden, R (2019) Science publishers review ethics of research on Chinese Minority Groups. Nature, 576, DOI: 10.1038/d41586-019-03775-y ↩
-
The Guardian (2019) US company to stop selling China equipment to build Uighur DNA database. Accessed 18 June 21. ↩
-
Erlich, Y (2018) Identity inference of genomic data using long-range familial searches. Science, 362(6415), DOI: 10.1126/science.aau4832 ↩ ↩2
-
Thomson, J (2019) The Effectiveness of Forensic Genealogy Techniques in the United Kingdom. Forensic Science International: Genetics , 7(1), DOI: 10.1016/j.fsigss.2019.10.169 ↩
-
Kaiser, J (2018) We will find you: DNA search used to nab Golden State Killer can home in on about 60% of white Americans. Science, DOI: 10.1126/science.aav7021 ↩
-
New York Times (2019) Your DNA Profile is Private? A Florida Judge Just Said Otherwise. Accessed 18 June 2021. ↩ ↩2
-
Kaiser, J (2019) A judge said police can search the DNA of 1 million Americans without their consent. What’s next?. Science, DOI: 10.1126/science.aba1428 ↩
-
FamilyTreeDNA (2021) FamilyTreeDNA Law Enforcement Guide. Accessed 18 June 2021. ↩
-
Brown, K (2019) Major DNA Testing Company Sharing Genetic Data With the FBI. Bloomberg. Accessed 18 June 2021. ↩
-
23andMe (2021) 23andMe Guide for Law Enforcement. Accessed 18 June 2021. ↩
-
Future of Privacy Forum (2018) Privacy Best Practices for Consumer Genetic Testing Services. Accessed 18 June 2021. ↩
-
UK Biobank (2007) UK Biobank Ethics and Governance Framework. Accessed 18 June 2021. ↩ ↩2
-
Genomics England (2020) Data in the Library. Accessed 18 June 2021. ↩
-
NHS Genomic Medicine Service (2020) Genomic Research and why it is important. Accessed 18 June 2021. ↩
-
O’Doherty, K (2016) If you build it, they will come: unintended future uses of organised health data collections. BMC Med Ethics, 17(1), DOI: 10.1186/s12910-016-0137-x ↩
-
Nature (2018) The ethics of catching criminals using their family’s DNA. Nature, 557(5), DOI: 10.1038/d41586-018-05029-9. ↩
-
Lenneman, B (2021) Enhancing phage therapy through synthetic biology and genome engineering. Current Opinion in Biotechnology, 68, DOI: 10.1016/j.copbio.2020.11.003 ↩
-
De Valle, I (2021) Translating New Synthetic Biology Advances for Biosensing Into the Earth and Environmental Sciences. Frontiers in Microbiology, 11, DOI: 10.3389/fmicb.2020.618373 ↩
-
Martinez, A (2018) Development of a GFP Fluorescent Bacterial Biosensor for the Detection and Quantification of Silver and Copper Ions. bioRxiv, 296079, DOI: 10.1101/296079 ↩
-
Charbonneau, M (2020) Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nature Communications, 11(1), DOI: 10.1038/s41467-020-15508-1 ↩
-
Cory, L (2014) ADXS-HPV: A therapeutic Listeria vaccination targeting cervical cancers expressing the HPV E7 antigen. Human Vaccines & Immunotherapeutics, 10(11), DOI: 10.4161/hv.34378 ↩
-
Shokravi, Z (2019) The Fourth-Generation Biofuel: A Systematic Review on Nearly Two Decades of Research from 2008 to 2019. Fossil Free Fuels, DOI: 10.1201/9780429327773-12 ↩
-
Cripwell, R (2019) Construction of industrial Saccharomyces cerevisiae strains for the efficient consolidated bioprocessing of raw starch. Biotechnology for Biofuels, 12(1), DOI: 10.1186/s13068-019-1541-5 ↩
-
François, J (2020) Synthetic Biology Applied to Carbon Conservative and Carbon Dioxide Recycling Pathways. Front. Bioeng. Biotechnol. 7(446), DOI: 10.3389/fbioe.2019.00446 ↩
-
Moradali, M (2020) Bacterial biopolymers: from pathogenesis to advanced materials. Nature Reviews Microbiology, 18(4), DOI: 10.1038/s41579-019-0313-3 ↩
-
de Lorenzo, V (2016) Bioremediation at a global scale: from the test tube to planet Earth. Microbial Biotechnology, 9(5), DOI: 10.1111/1751-7915.12399 ↩
-
Lorenzo, V (2018) The power of synthetic biology for bioproduction, remediation and pollution control. EMBO reports, 19(4), DOI: 10.15252/embr.201745658 ↩
-
Mora, R (2008) Enhanced bioremediation using whey powder for a trichloroethene plume in a high-sulfate, fractured granitic aquifer. Remediation Journal, 18(3), DOI: https://doi.org/10.1002/rem.20168 ↩
-
Hayes, B (2017) ‘Computing comes to life’ American Scientist, DOI: 10.1511/2001.22.204. Accessed 21 June 2021. ↩
-
Grozinger, L (2019) Pathways to cellular supremacy in biocomputing. Nature Communications, 10(1), DOI: 10.1038/s41467-019-13232-z ↩
-
Hutchison, C (2016) Design and synthesis of a minimal bacterial genome. Science, 351(6280), DOI: 10.1126/science.aad6253 ↩
-
Tachibana, C (2019) Beyond CRISPR: What’s current and upcoming in genome editing. Science, 365(6460), DOI: 10.1126/science.365.6460.1484-b ↩
-
Ostrov, N (2019) Technological challenges and milestones for writing genomes. Science, 366(6463), DOI: 10.1126/science.aay0339 ↩
-
Fredens, J (2019) Total synthesis of Escherichia coli with a recoded genome. Nature, 569, DOI: 10.1038/s41586-019-1192-5 ↩
-
Yeasts have several chromosomes, unlike bacteria. This added complexity means that yeast genome synthesis tends to be more difficult to achieve. ↩
-
A full collection of Nature publications on the project can be found here. Accessed 22 June 2021. ↩
-
Taking into account the 3.2 billion bases of the haploid human genome. A full diploid genome corresponds to 6.4 billion bases, and would cost $3.2 million. ↩
-
Ellis, T (2019) What is synthetic genomics anyway? The Biochemist, DOI: 10.1042/BIO04103006 ↩
-
Goldman, N (2013) Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature, 494, DOI: 10.1038/nature11875 ↩
-
Cumbers, J (2020) The Future Of Manufacturing Is Built With Biology. Or, How This Biotech Startup Is Challenging The Trillion-Dollar Global Chemical Industry. Forbes. Accessed 27 July 2021. ↩
-
For more information, see the Center of Excellence for Engineering Biology website and project page. Accessed 22 June 2021. ↩
-
Kuo, J (2018) Synthetic genome recoding: new genetic codes for new features. Current Genetics, 64, DOI: 10.1007/s00294-017-0754-z ↩
-
Willis, J (2018) Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nature Chemistry, 10, DOI: 10.1038/s41557-018-0052-5 ↩
-
DIYBiosphere (2018) ‘About’ DIYBiosphere. Accessed 21 June 2021. ↩
-
Biohackspace (2021) ‘London Biohackspace’ Biohackspace. Accessed 21 June 2021 ↩
-
SynbiCITE (2021) ‘About us’ SynbiCITE. Accessed 21 June 2021. ↩
-
Health and Safety Executive (2021) Genetically Modified Organisms (Contained Use) Regulations 2014 Public Register ↩
-
Wolinsky, H (2016) The FBI and biohackers: an unusual relationship. EMBO Reports, 17, DOI: 10.15252/embr.201642483 ↩
-
Hollis, A (2013) [Synthetic biology: ensuring the greatest global value.] (https://link.springer.com/article/10.1007/s11693-013-9115-5) Systems and Synthetic Biology, 7, DOI: 10.1007/s11693-013-9115-5 ↩
-
McKinsey Global Institute (2020) The Bio Revolution: Innovations transforming economies, societies, and our lives. McKinsey & Company. Accessed 18 June 2021. ↩
-
United Nations (2021) Global issues: population. Accessed 18 June 2021. ↩
-
Plumpton, H (2019) Climate Change and Agriculture. Parliamentary Office for Science and Technology. Accessed 28 July 2021. ↩
-
Tait-Burkard, C (2018) Livestock 2.0 – genome editing for fitter, healthier, and more productive farmed animals, Genome Biology, 19, DOI: 10.1186/s13059-018-1583-1 ↩ ↩2
-
Pardey, P (2014) A Bounds Analysis of World Food Futures: Global Agriculture Through to 2050. AARES, 58(4), DOI: 10.1111/1467-8489.12072 ↩
-
McPherron, A (1997) Double muscling in cattle due to mutations in the myostatin gene. PNAS, 94(23), DOI: 10.1073/pnas.94.23.12457 ↩
-
Piquerez, S (2014) Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front. Plant Sci., 5, DOI: 10.3389/fpls.2014.00671 ↩
-
Wray, N (2019) Complex Trait Prediction from Genome Data: Contrasting EBV in Livestock to PRS in Humans: Genomic Prediction. Genetics, 211(4), DOI: 10.1534/genetics.119.301859 ↩ ↩2
-
Brito, L (2020) Large-Scale Phenotyping of Livestock Welfare in Commercial Production Systems: A New Frontier in Animal Breeding. Front. Genet., 11, DOI: 10.3389/fgene.2020.00793 ↩
-
Georges, M (2018) Harnessing genomic information for livestock improvement. Nature Reviews Genetics, 20, DOI: 10.1038/s41576-018-0082-2 ↩
-
Romanov, M (2021) British Sheep Breeds as a Part of World Sheep Gene Pool Landscape: Looking into Genomic Applications. MDPI Animals, 11(4), DOI: 10.3390/ani11040994 ↩
-
Granleese, T (2015) Increased genetic gains in sheep, beef and dairy breeding programs from using female reproductive technologies combined with optimal contribution selection and genomic breeding values. Genetics Selection Evolution, 47, DOI: 10.1186/s12711-015-0151-3 ↩
-
Houston, R (2020) Harnessing genomics to fast-track genetic improvement in aquaculture. Nature Reviews Genetics, 21, DOI: 10.1038/s41576-020-0227-y ↩
-
Wang, X (2018) Genomic selection methods for crop improvement: current status and prospects. The Crop Journal, 6(4), DOI: 10.1016/j.cj.2018.03.001 ↩
-
Xu, Y (2020) Enhancing Genetic Gain through Genomic Selection: From Livestock to Plants. Plant Communications, 1(1), DOI: 10.1016/j.xplc.2019.100005 ↩
-
Meuwissen, T (2013) Accelerating Improvement of Livestock with Genomic Selection. Annual Review of Animal Biosciences, 1, DOI: 10.1146/annurev-animal-031412-103705 ↩ ↩2
-
Bhat, J (2016) Genomic Selection in the Era of Next Generation Sequencing for Complex Traits in Plant Breeding. Front. Genet., 7, DOI: 10.3389/fgene.2016.00221 ↩
-
Chu, J (2020) Suitability of Single-Nucleotide Polymorphism Arrays Versus Genotyping-By-Sequencing for Genebank Genomics in Wheat. Front. Plant Sci., 11, DOI: 10.3389/fpls.2020.00042 ↩
-
Wang, N (2020) Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Scientific Reports, 10, DOI: 10.1038/s41598-020-73321-8 ↩
-
Weigel, K (2012) Potential gains in lifetime net merit from genomic testing of cows, heifers, and calves on commercial dairy farms. Journal of Diary Science, 95(4), DOI: 10.3168/jds.2011-4877 ↩
-
Fraley, R (1983) Expression of bacterial genes in plant cells. PNAS, 80(15), DOI: 10.1073/pnas.80.15.4803 ↩
-
Hundleby, P (2018) Impacts of the EU GMO regulatory framework for plant genome editing. Food and Energy Security, 8(2), DOI: 10.1002/fes3.161 ↩
-
The Royal Society (2016) What GM crops are currently being grown and where? Accessed 18 June 2021. ↩
-
ISAAA (2018) Global Status of Commercialized Biotech/GM Crops in 2018: Biotech Crops Continue to Help Meet the Challenges of Increased Population and Climate Change. ISAAA Brief No. 54. Accessed 22 June 2021. ↩
-
World Health Organization (2014) Food, genetically modified. Accessed 18 June 2021. ↩
-
US Food and Drug Administration (2020) GMO Crops, Animal Food, and Beyond. Accessed 29 July 2021. ↩
-
International Rice Research Institute (2021) Philippines becomes first country to approve nutrient-enriched “Golden Rice” for planting. Accessed 29 July 2021. ↩
-
Dubock, A (2017) An overview of agriculture, nutrition and fortification, supplementation and biofortification: Golden Rice as an example for enhancing micronutrient intake. Agriculture & Food Security, 6(59), DOI: 10.1186/s40066-017-0135-3 ↩ ↩2
-
Owens, B (2018) Golden Rice is safe to eat, says FDA. Nat Biotechnol 36, DOI: 10.1038/nbt0718-559a ↩
-
Greenpeace International (2016) Nobel laureates sign letter on Greenpeace ‘Golden’ rice position – statement. Accessed 29 July 2021. ↩
-
Anderson, J (2019) Genetically Engineered Crops: Importance of Diversified Integrated Pest Management for Agricultural Sustainability. Front. Bioeng. Biotechnol. 7(24), DOI: 10.3389/fbioe.2019.00024 ↩
-
Department for Environment, Food, and Rural Affairs 2021) Genetic technologies regulation: government response. Accessed 29 September 2021. ↩ ↩2
-
Barnett, J (2016) Consumers’ confidence, reflections and response strategies following the horsemeat incident. Food Control, 59, DOI: 10.1016/j.foodcont.2015.06.021 ↩
-
Catalano, V (2016) Experimental review of DNA-based methods for wine traceability and development of a single-nucleotide polymorphism (SNP) genotyping assay for quantitative varietal authentication. J. Ag. & Food Chem., 64(37), DOI: 10.1021/acs.jafc.6b02560 ↩
-
Haynes, E (2019) The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food control, 101, DOI: 10.1016/j.foodcont.2019.02.010 ↩
-
Barcaccia, G (2016) DNA barcoding as a molecular tool to track down mislabelling and food piracy. MDPI Diversity, 8(1), DOI: 10.3390/d8010002 ↩
-
Carvalho, D (2017) Food metagenomics: Next generation sequencing identifies species mixtures and mislabelling within highly processed cod products. Food Control, 80, DOI: 10.1016/j.foodcont.2017.04.049 ↩
-
FERA (2019) Development of Metagenomic Methods for Determination of Origin - phase 2 - FA0160 Project Report. Accessed 28 July 2021. ↩
-
Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (2018) One Health Basics. Accessed 29 July 2021. ↩
-
Public Health England (2018) Implementing pathogen genomics. Accessed 29 July 2021. ↩
-
Grant, K (2018) What is Whole Genome Sequencing? PHE Blog. Accessed 29 July 2021. ↩
-
Early, R (2016) Global threats from invasive alien species in the twenty-first century and national response capacities. Nat Commun. 2016; 7, DOI: 10.1038/ncomms12485 ↩
-
Caffrey J (2014) Tackling invasive alien species in Europe: the top 20 issues. Management of Biological Invasions, 5(1), DOI: 10.3391/mbi.2014.5.1.01 ↩
-
Madden, M (2019) Using DNA barcoding to improve invasive pest identification at U.S. ports-of-entry. PLoS ONE, 14(9), DOI: 10.1371/journal.pone.0222291 ↩
-
Ratnasingham, S (2007) The Barcode of Life Data System. Mol. Ecol. Notes, 7(3), DOI: 10.1111/j.1471-8286.2007.01678.x ↩
-
Wasser, S (2007) Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban. Proceedings of the National Academy of Sciences. PNAS, 104(10), DOI: 10.1073/pnas.0609714104. ↩
-
Dormontt, E (2015) Forensic timber identification: It’s time to integrate disciplines to combat illegal logging. Biological Conservation, 191, DOI: 10.1016/j.biocon.2015.06.038 ↩
-
Lowe, A (2011) The Application of DNA methods to Timber Tracking and Origin Verification. IAWA Journal, 32(2), DOI: 10.1163/22941932-90000055 ↩ ↩2 ↩3
-
Lowe, A (2010) A DNA Method to Verify the Integrity of Timber Supply Chains; Confirming the Legal Sourcing of Merbau Timber From Logging Concession to Sawmill. Silvae Genetica, 59(1-6), DOI: 10.1515/sg-2010-0037 ↩
-
Double Helix (2019) Plant DNA evidence supports landmark Lacey Act conviction of BigLeaf Maple theft. Accessed 18 June 2021. ↩
-
Department of Justice, U.S. Attorney’s Office, Western District of Washington (2015) Tree Thieves and Mill Owner Indicted for Theft of Big Leaf Maples from National Forest. Accessed 18 June 2021. ↩
-
Forest Stewardship Council (2019) WorldForestID, Accessed 18 June 2021. ↩
-
Ong S (2013) Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16S rRNA amplicon sequences. PLoS ONE 8(4), DOI: 10.1371/journal.pone.0060811 ↩
-
Frantz, A (2006) [Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous] population.](https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1365-294X.2006.03022.x) Molecular Ecology, 15(11), DOI: 10.1111/j.1365-294X.2006.03022.x. ↩
-
Frantz, A (2004) Estimating population size by genotyping remotely plucked hair: the Eurasian badger. Journal of Applied Ecology, 41(5), DOI: 10.1111/j.0021-8901.2004.00951.x. ↩
-
Natural England (2019) Natural England’s Geoportal : England-wide data for great crested newts now available. Accessed 18 June 2021. ↩
-
Natural England (2019) Protecting great crested newts. Accessed 18 June 2021. ↩
-
Champer, J (2016) Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet 17, DOI: 10.1038/nrg.2015.34. ↩
-
Nash, A (2019) Integral gene drives for population replacement. Biol Open; 8(1): DOI: 10.1242/bio.037762. ↩
-
Faber, N (2021) Novel combination of CRISPR-based gene drives eliminates resistance and localises spread. Sci Rep, 11, DOI: 10.1038/s41598-021-83239-4. ↩
-
Oh, K (2021) Population genomics of invasive rodents on islands: Genetic consequences of colonization and prospects for localized synthetic gene drive. Evol Appl, 14, DOI: 10.1111/eva.13210. ↩
-
Kyrou, K (2018) A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol, 36, DOI: 10.1038/nbt.4245. ↩
-
DiCarlo, J (2015) Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol, 33, DOI: 10.1038/nbt.3412. ↩
-
Borevitz, J (2021) Utilizing genomics to understand and respond to global climate change. Genome Biology, 22, DOI: 10.1186/s13059-021-02317-y ↩
-
Waldvogel, A (2021) Evolutionary genomics can improve prediction of species’ responses to climate change. Evolution Letters, 4(1), DOI: 10.1002/evl3.154 ↩ ↩2
-
Waldvogel, A (2020) Climate Change Genomics Calls for Standardized Data Reporting. Front. Ecol. Evol., DOI: 10.3389/fevo.2020.00242 ↩
-
Gienapp, P (2008) Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol., 17(1), DOI: 10.1111/j.1365-294X.2007.03413.x ↩
-
Fitzpatrick, M (2014) Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecology Letters, 18(1), DOI: 10.1111/ele.12376 ↩
-
Bay, R (2018) Genomic signals of selection predict climate-driven population declines in a migratory bird. Science, 359(6371), DOI: 10.1126/science.aan4380 ↩
-
Exposito-Alonso, M (2017) Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nature Ecology & Evolution, 2, DOI: 10.1038/s41559-017-0423-0 ↩
-
Rellstab, C (2016) Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Molecular Ecology, 25(23), DOI: 10.1111/mec.13889 ↩
-
Stocks, J (2019) Genomic basis of European ash tree resistance to ash dieback fungus. Nat Ecol Evol, 3, DOI: 10.1038/s41559-019-1036-6. ↩
-
Harper, A (2016) Molecular markers for tolerance of European ash (Fraxinus excelsior) to dieback disease identified using Associative Transcriptomics. Sci Rep, 6, DOI: 10.1038/srep19335. ↩
-
Shi, J (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnology Journal, (15)2), DOI: 10.1111/pbi.12603 ↩
-
Adams, W (2021) Gene editing for climate: terraforming and biodiversity. Scottish Geographical Journal, 136(1-4), DOI: 10.1080/14702541.2020.1853869 ↩
-
United Nations Treaty Collection (2021) Cartagena Protocol on Biosafety to the Convention on Biological Diversity. Accessed 17 August 2021. ↩
-
Webber, B (2015) Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? PNAS, 112(34, DOI: 10.1073/pnas.1514258112 ↩
-
Haldane, J. B. S. (1938) Heredity and Politics. George Allen & Unwin Ltd, London. ↩
-
Singer, N (2018) Employees Jump at Genetic Testing. Is That a Good Thing? The New York Times, 15 April 2018. Accessed 18 June 2021. ↩
-
Lo, M (2017) Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nature genetics, 49(1), 152, DOI: 10.1038/ng.3736 ↩
-
Salminen, S (2018) Age, sex, and genetic and environmental effects on unintentional injuries in young and adult twins. Twin research and human genetics, 21(6), 502-506, DOI: 10.1017/thg.2018.61 ↩ ↩2
-
Kim, S (2018) Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PloS ONE, 13(7), DOI: 10.1371/journal.pone.0200785 ↩
-
Sulkava, S (2017) Common genetic variation near melatonin receptor 1A gene linked to job-related exhaustion in shift workers. Sleep, 40(1), DOI: 10.1093/sleep/zsw011 ↩
-
McKeown-Eyssen, G (2004) Case-control study of genotypes in multiple chemical sensitivity: CYP2D6, NAT1, NAT2, PON1, PON2 and MTHFR. International Journal of Epidemiology, 33(5), DOI: 10.1093/ije/dyh251 ↩
-
Genewatch UK (2003) Genetic Testing in the Workplace ↩
-
Mbemi, A (2020) Impact of Gene–Environment Interactions on Cancer Development. Environmental Research and Public Health, 17, DOI: 10.3390/ijerph17218089 ↩
-
Liu, C (2015) Genome-wide gene–asbestos exposure interaction association study identifies a common susceptibility variant on 22q13.31 associated with lung cancer risk. Cancer Epidemiology and Prevention Biomarkers, 1564-1573, 24(10), DOI: 10.1158/1055-9965.EPI-15-0021 ↩
-
In the USA, the Genetic Information Non-discrimination Act (GINA) made it illegal for employers to use an individual’s genetic information to determine decisions regarding hiring, promotion or firing. See Suter, S. M. (2018). GINA at 10 years: the battle over ‘genetic information’ continues in court. Journal of Law and the Biosciences, 495-526, 5(3), DOI: https://doi.org/10.1093/jlb/lsz002 ↩
-
There have been some successful challenges using GINA and some evidence that it is an effective deterrent of genetic discrimination in recruitment. See ASHG Perspective (2019). Prohibiting Genetic Discrimination to Promote Science, Health, and Fairness. Science Direct. The American Journal of Human Genetics 104, 6–7, DOI: https://doi.org/10.1016/j.ajhg.2018.12.005 ↩
-
Information Commissioner’s Office (ICO) (2011) The employment practices code. The ICO announced an update to the code in early 2021 that is currently under development. ↩
-
Clayton, E (2003). Ethical, legal, and social implications of genomic medicine. New England Journal of Medicine, 349(6), 562-569, DOI: 10.1056/NEJMra012577 ↩
-
Rudnik-Schöneborn, S (2012) Genetic tests in sports medicine-many studies, little impact. Genomics, Society and Policy, 8(1), DOI: https://doi.org/10.1186/1746-5354-8-1-13 ↩
-
Corrado, D (2016) Trends in Sudden Cardiovascular Death in Young Competitive Athletes After Implementation of a Preparticipation Screening Program. JAMA, 296(13), DOI: 10.1001/jama.296.13.1593 ↩
-
IAAF (2019) Eligibility Regulations for the Female Classification (Athletes with Differences of Sex Development). Accessed 22 June 2021. ↩
-
Wonkam, A (2010) Beyond the Caster Semenya Controversy: The Case of the Use of Genetics for Gender Testing in Sport. Journal of Genetic Counseling, 19(6), DOI: 10.1007/s10897-010-9320-2 ↩
-
Hercher, L (2010) Gender Verification: A Term Whose Time has Come and Gone. Journal of Genetic Counselling, 19(6), DOI: 10.1007/s10897-010-9323-z ↩
-
Varley, I (2008) ‘One club wants to use a gene-test to spot the new Ronaldo. Is this football’s future?’ The Guardian. Accessed 21 June 2021. ↩
-
Varley, I (2018) The current use, and opinions of elite athletes and support staff in relation to genetic testing in elite sport within the UK. Biology of Sport, 35, DOI: 10.5114/biolsport.2018.70747 ↩
-
Lemon J (2018) China will begin using genetic testing to select Olympic athletes. Newsweek. Accessed 8 June 2021. ↩
-
Synovitz & Eshanova (2014) Uzbekistan Is Using Genetic Testing to Find Future Olympians. The Atlantic, 6 February 2014. Accessed 18 June 2021. ↩
-
Webborn, N (2015) Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus. British Journal of Sports Medicine, 49(23), DOI: 10.1136/bjsports-2015-095343 ↩
-
Vlahovich, N (2017) Ethics of genetic testing and research in sport: a position statement from the Australian Institute of Sport. British journal of sports medicine, 51(1), 5-11, DOI: 10.1136/bjsports-2016-096661 ↩
-
Posthumus, M., & Collins, M. (Eds.), (2016) Genetics and sports. Karger Medical and Scientific Publishers. ↩
-
Pickering, C (2019) Can Genetic Testing Identify Talent for Sport? Genes, 10(12), 972, DOI: 10.3390/genes10120972 ↩ ↩2
-
Ahmetov, I (2015) Current progress in sports genomics. Advances in Clinical Chemistry (2015), 247-314, 70, DOI: 10.1016/bs.acc.2015.03.003 ↩
-
Priscilla, M (2005) ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. Journal of Applied Physiology, (2005), 154-163, 99(1), DOI: 10.1152/japplphysiol.01139.2004 ↩
-
Pickering, C (2017) ACTN3: More than Just a Gene for Speed. Frontiers in Physiology, 8, DOI: 10.3389/fphys.2017.01080 ↩
-
Amir, O (2007) The ACE deletion allele is associated with Israeli elite endurance athletes. Experimental Physiology, 92(5), DOI: 10.1113/expphysiol.2007.038711 ↩
-
Droma, Y (2008) Adaptation to High Altitude in Sherpas: Association with the Insertion/Deletion Polymorphism in the Angiotensin-Converting Enzyme Gene. Wilderness & Environmental Medicine, 19(1), DOI: 10.1580/06-WEME-OR-073.1 ↩
-
Voroshin, I (2008) Dependence of endurance performance on ACE gene polymorphism in athletes. Human Physiology, 34(1), DOI: 10.1134/S0362119708010180 ↩
-
De la Chappelle, A (1993) Truncated erythropoietin receptor causes dominantly inherited benign human erythrocytosis. Proceedings of the National Academy of Sciences, 90(10), DOI: 10.1073/pnas.90.10.4495 ↩
-
Rankinen, T (2016) No evidence of a common DNA variant profile specific to world class endurance athletes. PloS ONE, 11(1), DOI: 10.1371/journal.pone.0147330 ↩
-
Diaz Ramirez, J (2020) The GALNTL6 Gene rs558129 Polymorphism Is Associated with Power Performance. Journal of Strength and Conditioning Research, 34(11), DOI: 10.1519/JSC.0000000000003814 ↩
-
Al-Khelaifi, F (2019) Metabolic GWAS of elite athletes reveals novel genetically-influenced metabolites associated with athletic performance. Sci Rep 9, 19889, DOI: 10.1038/s41598-019-56496-7 ↩
-
Carrard, F (2011) Sports and politics on the international scene. Rivista Di Studi Politici Internazionali, 25-32, 78(1). ↩
-
Patel, S (2019) Exploring the Regulation of Genetic Testing in Sport. Entertainment and Sports Law Journal 17(1), DOI: 10.16997/eslj.223 ↩ ↩2
-
WADA (2021) International Standard Prohibited List 2021. Accessed 18 June 2021. ↩
-
Vernec, A (2014) The Athlete Biological Passport: an integral element of innovative strategies in antidoping. Br J Sports Med. 48(10), DOI: 10.1136/bjsports-2014-093560. ↩
-
WADA (2021) Guidelines - Gene Doping Detection Based on Polymerase Chain Reaction (PCR). Accessed 30 July 2021. ↩
-
Macur, J (2014) Fighting for the body she was born with. The New York Times. Accessed 14 June 2021. ↩
-
Imray, G (2021) Semenya taking case to European Court of Human Rights. ABC News. Accessed 14 June 2021. ↩
-
Brzeziańska, E (2014) Gene doping in sport–perspectives and risks. Biology of sport, 31(4), DOI: 10.5604/20831862.1120931. ↩
-
Camporesi, S (2016) Ethics, genetic testing, and athletic talent: children’s best interests, and the right to an open (athletic) future. Physiological genomics, 191-195, 48(3), DOI: 10.1152/physiolgenomics.00104.2015 ↩
-
Hebbar, R (2011) The Impact of the Genetic Information Nondiscrimination Act on Sports Employers: A Game of Balancing Money, Morality, and Privacy. Willamette Sports Law Journal, (2011), 52-64, 8(2). ↩
-
Wagner, J (2013) Playing with heart and soul… and genomes: sports implications and applications of personal genomics. PeerJ, 1, e120, DOI: 10.7717/peerj.120 ↩
-
Butala, B (2017) Muscular body build and male sex are independently associated with malignant hyperthermia susceptibility. Canadian Journal of Anesthesia, 64(4), DOI: 10.1007/s12630-017-0815-2 ↩
-
Rietveld, C (2013) GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science, 1467-1471, 340(6139), DOI: 10.1126/science.1235488 ↩
-
Department for Education and Skills (2004) A National Conversation about Personalised Learning. Accessed 18 June 2021. ↩
-
Sabatello, M (2021) A Genomically Informed Education System? Challenges for Behavioral Genetics. Journal of Law, Medicine & Ethics, 130-144, 46(1). https://doi.org/10.1177/1073110518766027 ↩
-
Branigan, A (2013) Variation in the Heritability of Educational Attainment: An International Meta-Analysis. Social Forces, 92(1), DOI: 10.1093/sf/sot076 ↩
-
Von Stumm, S (2020) Predicting educational achievement from genomic measures and socioeconomic status. Developmental Science, 23(3), DOI: 10.1111/desc.12925 ↩ ↩2
-
Rimfeld, K (2016) Genetics affects choice of academic subjects as well as achievement. Scientific Reports, 6(1), DOI: 10.1038/srep26373 ↩
-
Shakeshaft, N (2013) Strong Genetic Influence on a UK Nationwide Test of Educational Achievement at the End of Compulsory Education at Age 16. PLoS ONE, 8(12), DOI: 10.1371/journal.pone.0080341 ↩ ↩2
-
Sneikers, S (2017) Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nature Genetics, 49(7), DOI: 10.1038/ng.3869 ↩
-
Trampush, J (2017) GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Molecular Psychiatry, 22(3), DOI: 10.1038/mp.2016.244 ↩
-
Harden, K (2020) Genetic associations with mathematics tracking and persistence in secondary school. npj Science of Learning, 5(1), DOI: 10.1038/s41539-020-0060-2 ↩
-
Morris, T (2018) Testing the validity of value-added measures of educational progress with genetic data. British Educational Research Journal, 44(5), DOI: 10.1002/berj.3466 ↩
-
Allegrini, A (2019) Genomic prediction of cognitive traits in childhood and adolescence. Molecular Psychiatry, 24(6). DOI: 10.1038/s41380-019-0394-4 ↩
-
Gialluisi, A (2020) Genome-wide association study reveals new insights into the heritability and genetic correlates of developmental dyslexia. Molecular Psychiatry, DOI: https://doi.org/10.1038/s41380-020-00898-x ↩ ↩2 ↩3
-
Karavani, E (2019) Screening Human Embryos for Polygenic Traits Has Limited Utility. Cell, 179(6). DOI: 10.1016/j.cell.2019.10.033 ↩
-
Gialluisi, A (2019) Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia. Translational Psychiatry, 9(1), DOI: https://doi.org/10.1038/s41398-019-0402-0 ↩
-
Smith-Wooley, E (2018) Differences in exam performance between pupils attending selective and non-selective schools mirror the genetic differences between them. npj Science of Learning, 3(1), DOI: 10.1038/s41539-018-0019-8 ↩
-
Trejo, S (2018) Schools as Moderators of Genetic Associations with Life Course Attainments: Evidence from the WLS and Add Health. Sociological Science, 5, DOI: 10.15195/v5.a22 ↩
-
Cesarini, D (2017) Genetics and educational attainment. npj Science of Learning, 2(1), DOI: 10.1038/s41539-017-0005-6 ↩ ↩2
-
Dowling, K (2018) ‘Genetic discrimination in education: what’s the risk?’ Bill of Health. Accessed 18 June 2021. ↩
-
Mosier, H (1960) Sexually deviant behavior in Klinefelter’s syndrome. The Journal of Pediatrics, 57(3), DOI: 10.1016/s0022-3476(60)80256-9. ↩
-
Wakeling, A (1972) Comparative study of psychiatric patients with Klinefelter’s syndrome and hypogonadism. Psychological Medicine, 2(2), DOI: 10.1017/S0033291700040617 ↩
-
O’Donovan, R (2018) Klinefelter’s syndrome and sexual offending–a literature review. Criminal Behaviour and Mental Health, 28(2), DOI: 10.1002/cbm.2052 ↩
-
Stockholm, K (2012) Criminality in men with Klinefelter’s syndrome and XYY syndrome: a cohort study. BMJ Open, 2(1), DOI : 10.1136/bmjopen-2011-000650 ↩
-
Mason, D (1994) The heritability of antisocial behavior: A meta-analysis of twin and adoption studies. Journal of Psychopathology and Behavioral Assessment, 16(4), DOI: 10.1007/BF02239409 ↩
-
Tielbeek, J (2017) Genome-Wide Association Studies of a Broad Spectrum of Antisocial Behavior. JAMA Psychiatry, 74(12), DOI: 10.1001/jamapsychiatry.2017.3069 ↩
-
Which encodes for T-Cadherin, a neuronal membrane adhesion protein. Variants in this gene have been associated with poor impulse control and ADHD. ↩
-
Tiihonen, J (2015) Genetic background of extreme violent behavior. Molecular psychiatry, 20(6), DOI: 10.1038/mp.2014.130 ↩
-
Wertz, J (2018) Genetics and crime: Integrating new genomic discoveries into psychological research about antisocial behavior. Psychological science, 29(5), DOI: 10.1177/0956797617744542 ↩
-
Richmond-Rakerd, L (2020) A polygenic score for age‐at‐first‐birth predicts disinhibition. Journal of Child Psychology and Psychiatry, 61(12), DOI: 10.1111/jcpp.13224 ↩
-
Hasin, D (2013) DSM-5 Criteria for Substance Use Disorders: Recommendations and Rationale. American Journal of Psychiatry, 170(8), DOI: 10.1176/appi.ajp.2013.12060782 ↩
-
Jensen, K (2016) A review of genome-wide association studies of stimulant and opioid use disorders. Molecular neuropsychiatry, 2(1), DOI: 10.1159/000444755 ↩
-
Prom-Wormley, E (2017) The genetic epidemiology of substance use disorder: a review. Drug and alcohol dependence, 180, DOI: 10.1016/j.drugalcdep.2017.06.040 ↩
-
Kozak, K (2018) The neurobiology of impulsivity and substance use disorders: implications for treatment. Ann N Y Acad Sci, 1451(1), DOI: 10.1111/nyas.13977 ↩
-
Xu, K (2020) Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nature Communications, 11(1), DOI: 10.1038/s41467-020-18489-3 ↩
-
Kranzler, H (2019) Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature Communications, 10(1), DOI: 10.1038/s41467-019-09480-8 ↩
-
Sun, J (2020) A genome-wide association study of cocaine use disorder accounting for phenotypic heterogeneity and gene–environment interaction. Journal of Psychiatry and Neuroscience, 45(1), DOI: 10.1503/jpn.180098 ↩
-
Sun, Y (2021) Identification of novel risk loci with shared effects on alcoholism, heroin, and methamphetamine dependence. Molecular Psychiatry, 26(4), DOI: 10.1038/s41380-019-0497-y ↩
-
Sanchez-Roige, S (2019) Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. American Journal of Psychiatry, 176(2), DOI: 10.1176/appi.ajp.2018.18040369 ↩
-
McSwiggan, S (2017) The forensic use of behavioral genetics in criminal proceedings: Case of the MAOA-L genotype. International journal of law and psychiatry, 50, DOI: 10.1016/j.ijlp.2016.09.005 ↩
-
Scurich, N (2017) Behavioural genetics in criminal court. Nature human behaviour, 1(11), DOI: 10.1038/s41562-017-0212-4 ↩
-
European Commission (2019) Genome-wide association studies, polygenic scores and social science genetics: overview and policy implications. JRC Technical Reports. JRC F7 - Knowledge Health and Consumer Safety, DOI: 10.2760/948992 ↩
-
Perry, W (2013) Predictive Policing: The Role of Crime Forecasting in Law Enforcement Operations. RAND Corporation, DOI: 10.7249/RR233 ↩
-
Green, B (2017) Modeling Contagion Through Social Networks to Explain and Predict Gunshot Violence in Chicago, 2006 to 2014. JAMA Internal Medicine, 177(3), DOI: 10.1001/jamainternmed.2016.8245 ↩
-
Lau, T (2020) Predictive Policing Explained. Brennan Centre for Justice. Accessed 21 June 2021. ↩
-
HM Government, Association of British Insurers (2018) Code on Genetic Testing and Insurance. Accessed 21 June 2021. ↩ ↩2 ↩3
-
Widdows, H (2011) The Ethics of Biobanking: Key Issues and Controversies. Health Care Analysis, (2011), 19(3), DOI: 10.1007/s10728-011-0184-x ↩
-
Ipsos Mori, Genomics England, Sciencewise and UKRI (2019) A public dialogue on genomic medicine: time for a new social contract? Accessed 21 June 2021. ↩ ↩2 ↩3
-
Linnér, R (2019) Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nature Genetics, 51(2), 245-257, DOI: 10.1038/s41588-018-0309-3 ↩
-
Giuseppe Longo, U (2015) Unravelling the genetic susceptibility to develop ligament and tendon injuries. Current stem cell research & therapy, 10(1), 56-63, DOI: 10.2174/1574888x09666140710112535 ↩
-
Magnusson, K (2021) High genetic contribution to anterior cruciate ligament rupture: Heritability ~69%. British Journal of Sports Medicine, 55(7), DOI: 10.1136/bjsports-2020-102392 ↩
-
Handel, B (2013) Adverse selection and inertia in health insurance markets. American Economic Review 103(7), DOI: 10.1257/aer.103.7.2643 ↩
-
Weyman, M (2019) Swiss RE SONAR 2019: New emerging risk insights, Swiss Re Institute, pages 36-37. Accessed 21 June 2021. ↩
-
Macdonald, A (2011) The impact of genetic information on the insurance industry: conclusions from the ‘bottom-up’ modelling programme. ASTIN Bulletin: The Journal of the IAA, 343-376, 41(2), DOI: 10.2143/AST.41.2.2136981 ↩
-
The Geneva Association (2017) Genetics and Life Insurance: A View Into the Microscope of Regulation Accessed 21 June 2021. ↩
-
Note: this table amends the original GINA 5 categories into 6, as we felt that the USA position warranted a separate category. ↩
-
House of Commons Science and Technology Committee (2001) Science and Technology – fifth report. Accessed 21 June 2021. ↩
-
HM Government & Association of British Insurers (2005) Concordat and Moratorium on Genetics and Insurance. Accessed 21 June 2021. ↩
-
Government Actuaries’ Department (2018) Genetic Testing: Insurance and the NHS. e-news from GAD, Issue 34. Accessed 21 June 2021. ↩
-
Godard, B (2003) Genetic information and testing in insurance and employment. European Journal of Human Genetics, 11(Suppl. 2), DOI: 10.1038/sj.ejhg.5201117 ↩
-
Suther, S (2009) Barriers to the use of genetic testing: a study of racial and ethnic disparities. Genetics in Medicine, 655-662, 11(9), DOI: 10.1097/GIM.0b013e3181ab22aa ↩
-
Nye, C (2018) ‘Biohacker: Meet the people ‘hacking’ their bodies’ BBC News. Accessed 21 June 2021. ↩
-
Esrick, E (2021) Post-Transcriptional Genetic Silencing of BCL11A to Treat Sickle Cell Disease. New England Journal of Medicine, 384(3), DOI: 10.1056/NEJMoa2029392 ↩
-
Frangoul, H (2021) CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. New England Journal of Medicine, 384(3), DOI: 10.1056/NEJMoa2031054 ↩
-
Stadtmauer, E (2020) CRISPR-engineered T cells in patients with refractory cancer. Science, 367(6481), DOI: 10.1126/science.aba7365 ↩
-
Cyranoski, D (2018) Genome-edited baby claim provokes international outcry. Nature, 563, DOI: 10.1038/d41586-018-07545-0 ↩
-
Cyranoski, D (2018) CRISPR-baby scientist fails to satisfy critics. Nature, 564, DOI: 10.1038/d41586-018-07573-w ↩
-
Joy, M (2019) CCR5 Is a Therapeutic Target for Recovery after Stroke and Traumatic Brain Injury. Cell, 176(5), DOI: 10.1016/j.cell.2019.01.044 ↩
-
Regalado, A (2019) China’s CRISPR twins might have had their brains inadvertently enhanced. MIT Technology Review. Accessed 22 June 2021. ↩
-
Reardon, S (2019) Gene edits to ‘CRISPR babies’ might have shortened their life expectancy. Nature, 570, DOI: 10.1038/d41586-019-01739-w ↩
-
Sample, I (2019) Chinese scientist who edited babies’ genes jailed for three years. The Guardian. Accessed 22 June 2021. ↩
-
Example product: Amino Labs (2021) ‘About us’ Amino labs. Accessed 21 June 2021. ↩
-
Robertson, S (2019) ‘California clamps down on amateur use of gene-editing technologies’ News-Medical. Accessed 21 June 2021. ↩
-
Lussenhop, J (2017) ‘Why I injected myself with an untested gene therapy’ BBC News. Accessed 21 June 2021. ↩
-
Cornish, C (2018) ‘The biohackers trying to alter their DNA at home’ Financial Times. Accessed 21 June 2021. ↩
-
US Food and Drug Administration (2017) ‘Information About Self-Administration of Gene Therapy’ FDA. Accessed 21 June 2021. ↩
-
Associated Press (2016) ‘IOC says new gene doping test won’t be used in Rio’ ESPN. Accessed 21 June 2021. ↩
-
Everts, S (2016) ‘Athletes at Rio Olympics Face Advanced Antidoping Technology’ Chemical & Engineering News. Accessed 21 June 2021. ↩
-
Le Page, M (2017) ‘Anti-doping agency to ban all gene editing in sport from 2018’ New Scientist. Accessed 21 June 2021. ↩
-
UK Ministry of Defence (2021) Human Augmentation – The Dawn of a New Paradigm. Accessed 29 July 2021. ↩
-
Genomics England (2021) Participant Panel. Accessed 30 July 2021. ↩
-
Gibbons, S (2007) Are UK genetic databases governed adequately? A comparative legal analysis. Legal Studies, 27(2), DOI: 10.1111/j.1748-121X.2007.00045.x ↩ ↩2
-
Berger, K (2019) National and Transnational Security Implications of Asymmetric Access to and Use of Biological Data. Frontiers in Bioengineering and Biotechnology, 7, DOI: 10.3389/fbioe.2019.00021 ↩
-
UK Government (2021) National Security and Investment Act. Accessed 22 July 2021. ↩
-
Information Commissioner’s Office (2012) Anonymisation: managing data protection risk code of practice. Accessed 22 June 2021. ↩
-
Data Protection Act 2018, c. 12. Accessed 22 June 2021. ↩
-
General Data Protection Regulation (2016) Regulation (EU) 2016/679, Official Journal of the European Union, L119. Accessed 22 June 2021. ↩
-
Information Commissioner’s Office (2018) Guide to data protection. Accessed 22 June 2021. ↩
-
Selita, F (2020) Justice in the genomic and digital era: a ‘different world’ requiring ‘different law’. Legal Issues Journal, 8(1), ISSN 2516-1210 ↩
-
Ohm, P (2010) Broken promises of privacy: responding to the surprising failure of anonymization. UCLA Law Review, (2010), 1701-1778, 57(6). ↩
-
Office of the Victorian Information Commissioner (2018) Disclosure of myki travel information: Investigation under section 8C(2)(e) of the Privacy and Data Protection Act 2014 (Vic). ↩
-
Rothstein, M (2010) Is Deidentification Sufficient to Protect Health Privacy in Research? The American Journal of Bioethics, 10(9), DOI: 10.1080/15265161.2010.494215 ↩
-
Loukides, G (2010) The disclosure of diagnosis codes can breach research participants’ privacy. Journal of the American Medical Informatics Association, 17(3), DOI: 10.1136/jamia.2009.002725. ↩
-
Government Office for Science (2020) The future of citizen data systems. Accessed 22 June 2021. ↩ ↩2
-
Barwell, J (2018) Challenges in implementing genomic medicine: the 100,000 Genomes Project. Journal of Translational Genetics and Genomics, 2(13), DOI: 10.20517/jtgg.2018.17 ↩
-
Genomics England (2020) The Genomics England Research Environment. Accessed 22 June 2021. ↩
-
Nuffield Council on Bioethics (2015) The collection, linking and use of data in biomedical research and health care: ethical issues. Accessed 22 June 2021. ↩
-
Jones, K (2017) The SAIL Databank: 10 years of spearheading data privacy and research utility, 2007-2017. Sail Databank. Accessed 22 June 2021. ↩
-
Rahimzadeh, V (2016) An International Framework for Data Sharing: Moving Forward with the Global Alliance for Genomics and Health. Biopreservation and Biobanking, 14(3), DOI: 10.1089/bio.2016.0005 ↩
-
Knoppers, B (2014) Framework for responsible sharing of genomic and health-related data. The HUGO Journal, 8(1), DOI: 10.1186/s11568-014-0003-1 ↩
-
Repeated queries could allow data to be partially reconstructed. ↩
-
Royal Society (2019) Protecting privacy in practice: the current use, development and limits of privacy enhancing technologies in data analysis. Accessed 22 June 2021. ↩
-
Mittos, A (2017) Systematizing Genome Privacy Research: A Privacy-Enhancing Technologies Perspective. Proceedings on Privacy Enhancing Technologies (PoPETs), DOI: 10.2478/popets-2019-0006 ↩
-
A form of encryption that permits computations to be carried out on encrypted data without decrypting it. ↩
-
Fontaine, C (2007) A Survey of Homomorphic Encryption for Nonspecialists. EURASIP Journal on Information Security, DOI: https://doi.org/10.1155/2007/13801 ↩
-
Wood, A (2018) Differential privacy: a primer for a non-technical audience. Vanderbilt Journal of Entertainment & Technology Law, 21(17), DOI: 10.2139/ssrn.3338027 ↩
-
Hall, B (2012) Recent Research on the Economics of Patents. Annual Review of Economics, 4(1), DOI: 10.1146/annurev-economics-080511-111008 ↩
-
Sherkow, J (2015) The History of Patenting Genetic Material. Annual Review of Genetics, 49, DOI: 10.1146/annurev-genet-112414-054731 ↩ ↩2
-
Patenting ESTs: is it worth it? (1999) Nature Genetics, 21(2), DOI: 10.1038/5920 ↩
-
Cook-Deegan, R (2010) Patents in Genomics and Human Genetics. Annual Review of Genomics and Human Genetics, 11(1), DOI: 10.1146/annurev-genom-082509-141811 ↩
-
The legal protection of biotechnological inventions (1998) Directive 98/44/EC. Official Journal of the European Communities, L213/13. Accessed 22 June 2021. ↩
-
Cartwright-Smith, L (2014) Patenting Genes: What Does Association for Molecular Pathology V. Myriad Genetics Mean for Genetic Testing and Research? Public Health Reports, 129(3), DOI: 10.1177/003335491412900311 ↩
-
Cho, M (2003) Effects of Patents and Licenses on the Provision of Clinical Genetic Testing Services. The Journal of Molecular Diagnostics, 5(1), DOI: 10.1016/S1525-1578(10)60444-8 ↩
-
Servick, K (2019) Controversial U.S. bill would lift Supreme Court ban on patenting human genes. Science, DOI: 10.1126/science.aay2710 ↩
-
Sherkow, J (2017) CRISPR, Patents, and the Public Health. Yale Journal of Biology and Medicine, 90. ↩
-
The Understanding Patient Data project by the Wellcome Trust focuses on how to make uses of patient data more visible, understandable and trustworthy, for patients, the public and health professionals, and includes FAQs and case studies. ↩
-
Middleton, A (2020) Global Public Perceptions of Genomic Data Sharing: What Shapes the Willingness to Donate DNA and Health Data? The American Journal of Human Genetics, 107(4), DOI: 10.1016/j.ajhg.2020.08.023 ↩
-
Data Justice Lab. Advancing civic participation in algorithmic decision-making. Accessed 22 July 2021. ↩
-
Ada Lovelace Institute. The Ada Lovelace Institute supports Wellcome Trust to undertake citizen juries on fair data sharing in the NHS. Accessed 22 July 2021. ↩
-
Reynolds, J (2019) DNA testing at birth will be available for everyone. The Times. Accessed 22 June 2021. ↩
-
Genetic Alliance UK (2019) Fixing the Present, Building for the Future: Newborn Screening for Rare Conditions. Accessed 22 June 2021. ↩
-
Nuffield Council on Bioethics (2018) Bioethics briefing note: whole genome sequencing of babies. Accessed 22 June 2021. ↩
-
Goldenberg, A (2014) Parents’ interest in whole-genome sequencing of newborns. Genetics in Medicine, 16(1), DOI: 10.1038/gim.2013.76 ↩
-
Bombard, Y (2014). Public views on participating in newborn screening using genome sequencing. European Journal of Human Genetics, 22(11), DOI: 10.1038/ejhg.2014.22 ↩ ↩2
-
Machado, H (2019) What influences public views on forensic DNA testing in the criminal field? A scoping review of quantitative evidence. Human Genomics, 13(1), DOI: 10.1186/s40246-019-0207-5 ↩ ↩2 ↩3
-
Machado, H (2014) “Would you accept having your DNA profile inserted in the National Forensic DNA database? Why?” Results of a questionnaire applied in Portugal. Forensic Science International: Genetics, 8(1), DOI: 10.1016/j.fsigen.2013.08.014 ↩
-
Gamero, J (2008) A study of Spanish attitudes regarding the custody and use of forensic DNA databases. Forensic Science International: Genetics, 2(2), DOI: 10.1016/j.fsigen.2007.10.201 ↩
-
Guerrini, C (2018) Should police have access to genetic genealogy databases? Capturing the Golden State Killer and other criminals using a controversial new forensic technique. PLoS Biology, 16(10), DOI: 10.1371/journal.pbio.2006906 ↩
-
Zieger, M (2015) About DNA databasing and investigative genetic analysis of externally visible characteristics: A public survey. Forensic Science International: Genetics, 17, DOI: 10.1016/j.fsigen.2015.05.010 ↩
-
House of Lords Science and Technology Select Committee (2019) Forensic science and the criminal justice system: a blueprint for change - third report. Accessed 22 June 2021. ↩
-
Smith, M (2018) Majority of Brits support introducing ID cards. YouGov. Accessed 22 June 2021. ↩
-
Perrin, A (2020) About half of Americans are OK with DNA testing companies sharing user data with law enforcement. Pew Research Centre. Accessed 22 June 2021. ↩
-
Ibbetson, C (2020) Should police have access to private DNA data. YouGov. Accessed 22 June 2021. ↩
-
Plan France Médecine Génomique 2025 (2019) ‘[Project Homepage]’(https://pfmg2025.aviesan.fr/en/). Viewed 13 May 2021 ↩
-
Coyle, D (2018) ‘Genomics research: Chinese group to lead $400m investment’. The Irish Times. Viewed 13 May 2021 ↩
-
Ireland Strategic Investment Fund (2016) ‘Genomics Medicine Ireland raises $40M in Series A funding’. Viewed 13 May 2021 ↩
-
Ireland Strategic Investment Fund (2018) ‘400 Million Investment Programme Positions Ireland for Global Leadership in Genomic Research and Advanced Life Sciences’. Viewed 13 May 2021 ↩
-
Genuity Science (2020) ‘The Genuity Science Rare Disease Programme’. Viewed 13 May 2021 ↩
-
National Institutes of Health (2019) ‘All of Us Research Program Expands Data Collection Efforts with Fitbit’. Viewed 13 May 2021 ↩
