Shiga toxin-producing Escherichia coli (STEC) data: 2019
Updated 30 July 2025
Main points for 2019
The main points of the 2019 report are:
1. A total of 539 confirmed cases of Shiga toxin-producing Echerichia coli (STEC) O157 were reported in England and Wales in 2019.
2. The lowest incidence of STEC O157 was in the East Midlands region (0.56 per 100,000 population) and the highest in the Yorkshire and Humber region (1.51 per 100,000 population).
3. Children aged 1 to 4 years had the highest incidence of infection (3.28 per 100,000 population, CI 95% 2.63–4.04).
4. Nearly one-third of confirmed STEC O157 cases in England were hospitalised and 3% were reported to have developed haemolytic ureamic syndrome (HUS).
5. In England and Wales, detection of non-O157 STEC increased in line with the growing number of NHS labs implementing gastrointestinal (GI) diagnostics using polymerase chain reaction (PCR); in 2019, 768 culture-positive non-O157 STEC cases (655 in England, 113 in Wales) were reported.
6. A further 347 specimens in England and 66 in Wales were positive for Shiga toxins (stx) genes on PCR at the Gastrointestinal Bacteria Reference Unit (GBRU) but an organism was not cultured.
7. The most commonly isolated non-O157 STEC serogroup was STEC O26 (England: n=109/655, 17% and Wales: n=28/113, 25%).
8. Five outbreaks of STEC involving 65 cases in England were investigated in 2019.
Background
STEC, also known as Vero cytotoxin-producing Escherichia coli (VTEC), are bacteria that can cause gastroenteritis. Symptoms vary from mild to bloody diarrhoea and, in severe cases, can cause HUS, a serious and life threatening condition predominantly affecting the kidneys. A small proportion of patients, mainly children, develop HUS (1).
The main reservoir for STEC is cattle although it is also carried by other ruminants such as sheep, goats and deer. Transmission can occur through direct or indirect contact with animals or their environments, consumption of contaminated food or water, and person-to-person spread.
STEC infections can present as sporadic cases or as outbreaks. Large national and multinational outbreaks have been associated with foodborne transmission (2 to 4).
The most common serogroup of STEC causing illness in England is O157 (5). Other serogroups (termed non-O157) can also cause illness and have been implicated in outbreaks in England and elsewhere.
Frontline laboratories in England use culture methods to detect STEC O157 by its inability to ferment sorbitol on selective media (MacConkey agar). However, non-O157 STEC ferment sorbitol and there is no culture method to differentiate non-O157 STEC from non-pathogenic E.coli in frontline laboratories. Therefore, detection of non-O157 STEC relies on PCR.
The implementation and roll-out of a GI PCR at frontline hospital laboratories began in December 2013, and to December 2019 around 20% of frontline laboratories were using it. As a consequence, there has been a substantial increase in the detection of non-O157 cases. However, PCR is not universally used for detection of non-O157 STEC, and the true incidence remains unknown.
While non-O157 STEC can cause serious illness, variation exists among non-O157 STEC serogroups in their associations with severe disease, likely explained by differences in the virulence factors produced by different strains. STEC can produce 2 Shiga toxins (Stx), Stx1 (of which there are 4 subtypes 1a–1d) and/or Stx2 (of which there are 7 subtypes stx2a–2g).
The presence of Stx2, specifically subtype stx2a, is more likely to cause HUS (1), (6). The increasing numbers of non-O157 STEC has led to the need to prioritise the public health actions due to insufficient resources to follow up all cases.
Risk assessment, based on clinical symptoms and risk group of the patient and potential pathogenicity of the strain of STEC infecting the patient, is challenging. In response, new Interim Public Health Operational Guidance for STEC for the public health management of O157 and non-O157 STEC cases (including an algorithm to assist in follow-up decision making) was published by the STEC Guidelines Update Working Group in August 2018.
National enhanced surveillance of STEC in England and Wales has been undertaken since 2009. This report summarises the epidemiological data on confirmed cases of STEC O157 and non-O157 STEC cases in England in 2019 and compares it to previous years.
Methods
The National Enhanced Surveillance System for STEC (NESSS) infection in England began in January 2009 in order to supplement our understanding of the epidemiology of STEC infection. The system collects a standard data set of clinical, epidemiological and microbiological data for all STEC cases, in order to improve outbreak recognition and facilitate public health investigations. The data is collected from enhanced surveillance questionnaires (ESQs) and reconciled with laboratory reports associated with cases.
STEC is notifiable under the Public Health (Control of Diseases) Act 1984 and the Health Protection (Notification) Regulations 2010. In England, local diagnostic laboratories report presumptive cases of STEC to the UK Health Security Agency Health Protection Teams (UKHSA HPTs) and then refer samples to the GBRU for confirmation and further testing. Each HPT arranges for the ESQ to be completed for all cases to obtain a detailed history for the 7 days prior to onset of illness.
The ESQ collects:
- demographic details
- risk status
- clinical conditions
- exposures including travel, food and water consumption
- environmental exposures
- outbreak status
Completed ESQs are submitted to the national Gastrointestinal Infections team at UKHSA to be included in NESSS.
Data included in this report was validated and extracted from NESSS and cases meeting the case definitions below were included in analyses. Laboratory data for cases in Wales was extracted and validated from the UKHSA Gastro Data Warehouse (GDW). Welsh data is included in Figure 1 as only laboratory data was available and is excluded from other sections of the report.
Data from the 2019 Office for National Statistics (ONS) mid-year population estimates was used to provide denominators for the calculation of incidence rates. All dates for the figures are based on the receipt date of a sample specimen at the GBRU.
Both microbiological and epidemiological case definitions are described in tables 1 and 2 below.
Table 1: Microbiological case definitions
| Classification | Definition |
|---|---|
| Confirmed | Positive STEC culture or PCR positive for stx genes confirmed by GBRU |
| Probable | Suspect case with serum antibodies to lipopolysaccharide of E. coli O157 or other STEC, detected at the GBRU |
Table 2: Epidemiological case definitions
| Classification | Definition |
|---|---|
| Primary | A symptomatic case with no history of close contact with a confirmed case in the 7 days prior to onset of illness |
| Secondary | Case with a date of onset more than 4 days after the primary case or where transmission is believed to be through exposure to a primary case |
| Unsure | It is not possible to determine whether the case is primary or secondary with the information available. This may be because the patient was lost to follow-up, is asymptomatic or in an outbreak where it is not possible to identify the primary case(s) |
| Travel-associated | Case who has reported any travel outside of the UK in the 7 days prior to their date of onset of illness |
| Asymptomatic | A person from whom STEC was identified through contact screening procedures but who is asymptomatic |
Data
Cases of STEC in England and Wales in 2019
In 2019, 1,720 confirmed cases of STEC were reported in England and Wales; these comprised 539 culture-confirmed cases of STEC serogroup O157 (515 cases in England and 24 in Wales) and 768 cases (655 in England, 113 in Wales) where a serogroup other than O157 was isolated (non-O157). For a further 413 cases, samples were confirmed as STEC by testing positive by PCR for stx genes, but STEC was not cultured (347 in England, 66 in Wales).
Five confirmed cases were infected with multiple serogroups:
- O157 and O26
- O26 and O103
- O76 and O113
- O91 and O128ab
- O146 and O91
There were 13 probable cases with serological evidence of STEC infection, with antibodies detected to O157 lipopolysaccharides in 11 cases (England: 10, Wales: 1), for O111 lipopolysaccharides in one case, and for O26 lipopolysaccharides in another case.
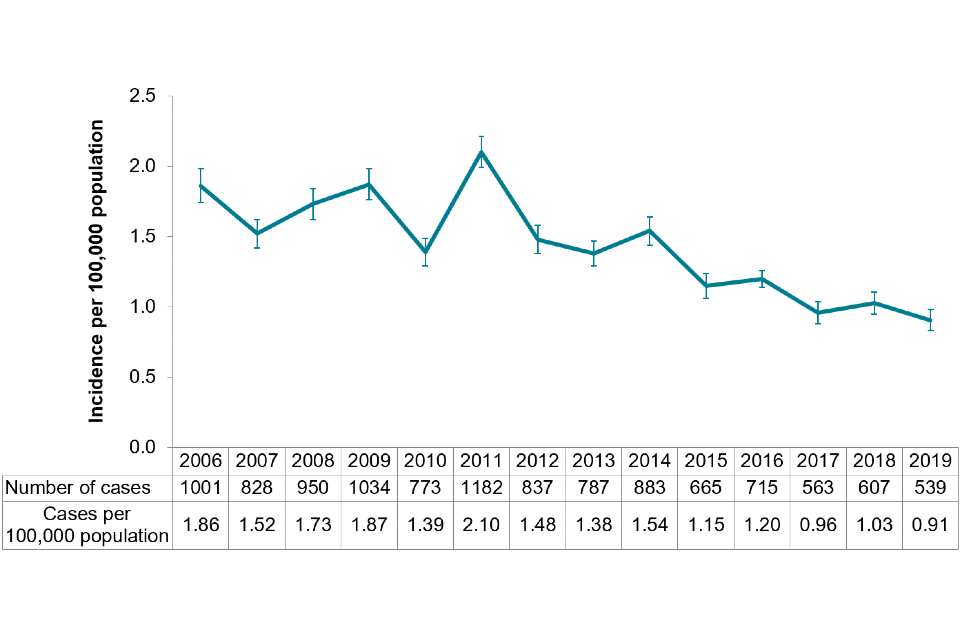
The crude incidence rate of confirmed STEC O157 in England and Wales was 0.91 per 100,000 cases (95% CI 0.83–0.99), continuing the downward trend observed since 2015 (Figure 1). It is the lowest number of cases reported annually since 1996, when testing began in England for STEC O157 on all faecal specimens from patients with suspected gastrointestinal infection (7).
Figure 1: Incidence of Shiga toxin producing Escherichia coli (STEC) O157 culture-confirmed cases by year, England and Wales, 2006 to 2019
Cases of STEC in 2019 in England
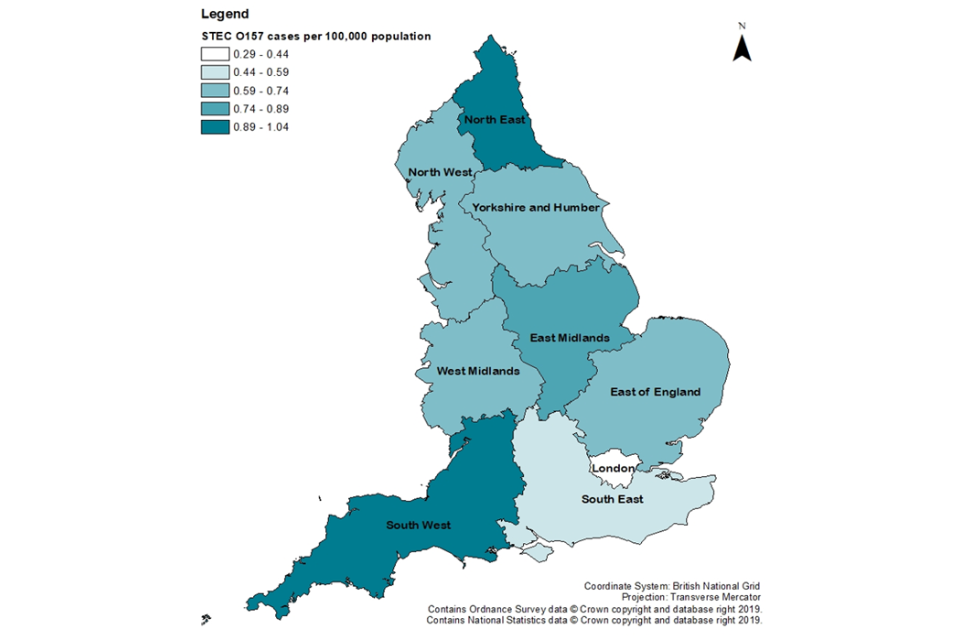
Figure 2: Incidence of STEC O157 in England by region, 2019 only
Coordinate System: British National Grid
Projection: Transverse Mercator
Contains Ordinance Survey data © copyright and database right 2019
Contains National Statistics data © copyright and database right 2019
The highest incidence of STEC O157 was in Yorkshire and Humber (1.51 cases per 100,000 population, 95% CI 1.20–1.87) and the lowest was in the East Midlands (0.56 cases per 100,000 population, 95% CI 0.37–0.81) (Figure 2).
Age, gender and seasonality of STEC O157 cases
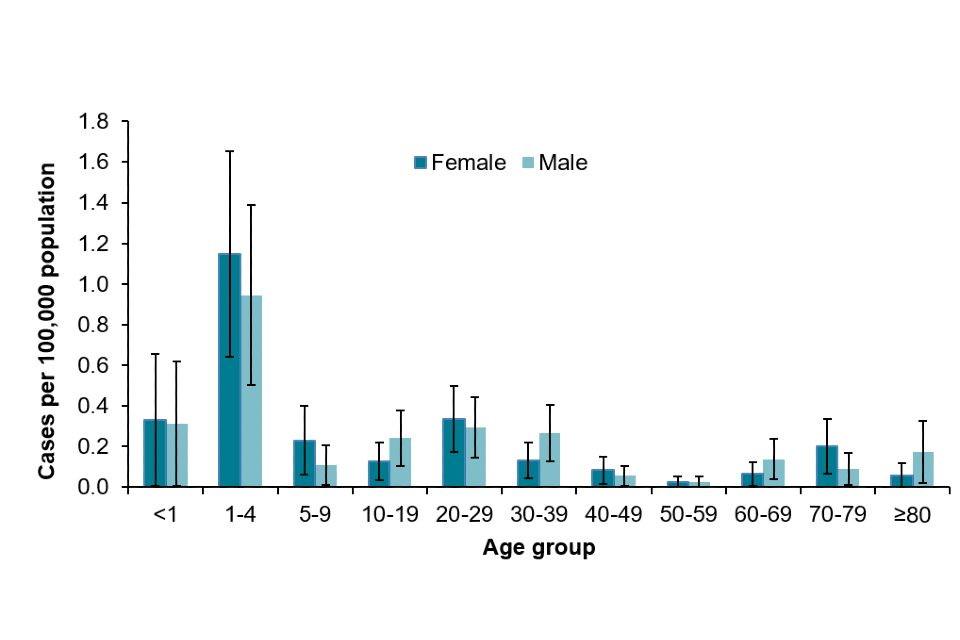
Of 515 confirmed STEC O157 cases in England, 280 (54%) were female. Children aged 1 to 4 years had the highest incidence of infection (3.28 per 100,000 population, CI 95% 2.63–4.04). Overall, females had a higher incidence across all age groups, with the exception of those between 1 to 4 years of age, and 10 to 19 years of age (Figure 3).
Figure 3: Age-specific incidence rate of STEC O157 cases in England, 2019 only
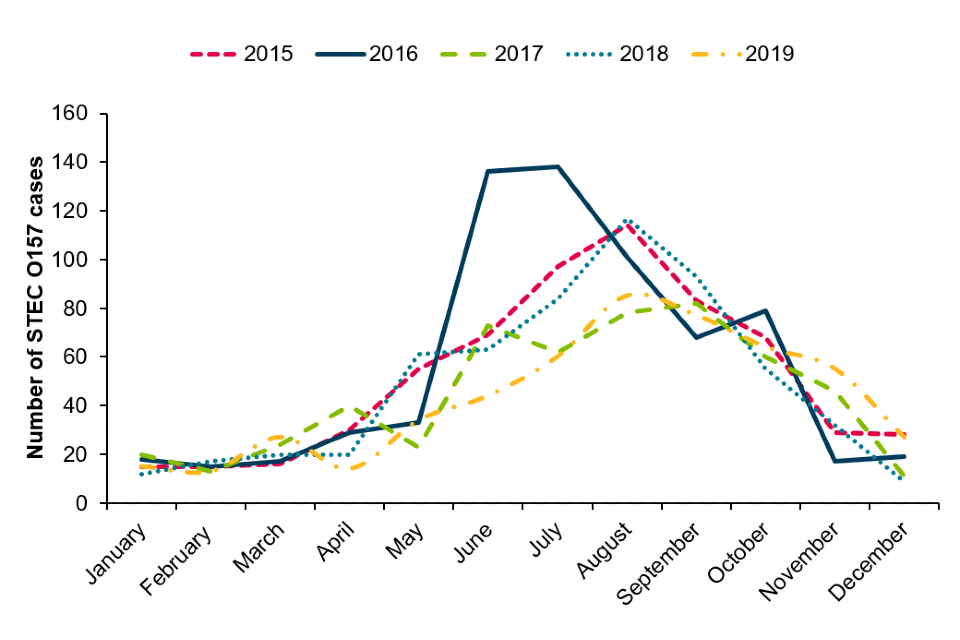
As per previous years, STEC O157 infections displayed a distinct seasonality with the peak of infection in the summer months (Figure 4).
Figure 4: Seasonal trend of laboratory confirmed cases of STEC O157 in England, 2015 to 2019
Severity of illness
Of the 515 confirmed STEC O157 cases in England in 2019, ESQs were received for 500 (97%). Of those, symptoms were reported for 482 (96%) cases, the majority of which reported diarrhoea (93%, n=467) as well as bloody stools (58%, n=290).
Other symptoms included abdominal pain (402, 80%), nausea (236, 47%), vomiting (150, 30%), and fever (144, 29%). Hospitalisation occurred in 29% (n=147) of cases; duration of hospitalisation ranged from 1 to 10 days with median stay of 2 days.
During 2019, HUS occurred in 13 confirmed (3%) and 5 probable STEC O157 cases. Nearly one-quarter of HUS cases were under 5 years of age (n=4, 22%) with a median age of 13 (range 1 to 75 years old). The incidence of STEC O157 HUS in children under 5 in 2019 was 0.12 per 100,000 cases per population (CI 0.03–0.31). No deaths were reported among confirmed or probable STEC O157 cases.
Transmission routes
In England, of the 500 confirmed cases for whom ESQs were received during 2019, there were 453 primary or co-primary cases, 29 secondary cases, and 18 asymptomatic contacts. One hundred and fifty-four cases (31%) were travel-related, and of those, 48 spent their entire incubation period (7 days prior to onset) abroad. The most frequently reported travel destinations included Turkey, Egypt, and Cyprus.
Frequently reported subtypes of STEC O157
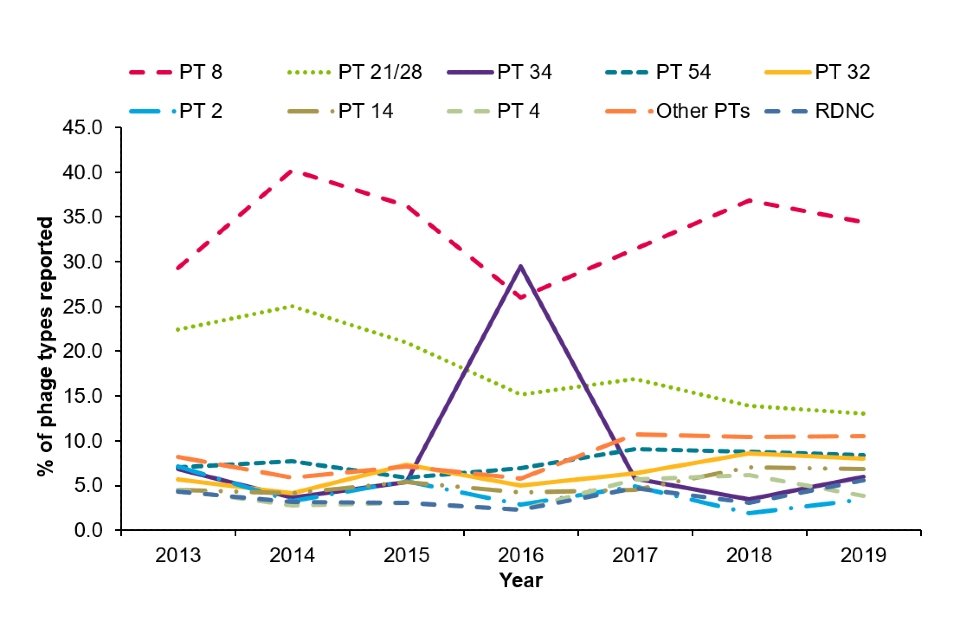
As with previous years, phage type (PT) 8 and 21/28 were the most commonly reported STEC O157 phage types in 2019 (Figure 5), with a combined proportion of 47%. These were followed by PT 54 (8%), PT 32 (8%) and PT 14 (7%). The peak in PT34 in 2016 was due to a large outbreak linked to mixed-salad leaves, where 165 cases were reported between 31 May and 29 July 2016 (4).
Figure 5: Number of STEC O157 cases by the 10 of the most common phage types, 2013 to 2019
Among 515 isolates from confirmed STEC O157 cases in England in 2019, most (n=322, 62%) had Stx2 shiga-toxin only, 37% (n=190) had Stx1+2, and 1% (n=3) had Stx1 only. Of those isolates with Stx2 or Stx1+2, 41% (n=209) had stx2a; the subtype most likely to cause HUS. All 515 isolates had the eae gene.
Non-O157 STEC cases in England and Wales
Historically, cases of non-O157 STEC have been under ascertained, with 89 cases of STEC non-O157 reported between 2009 and 2013, prior to PCR being implemented.
Following the increase in recent years in frontline laboratories using PCR, there has been a significant increase in the detection of non-O157 STEC in England. It is not possible to estimate a denominator for incidence calculations for non-O157 STEC because details of contract arrangements for referral of samples from primary care and catchment areas of each diagnostic laboratory using PCR are not known.
In 2019, of 5,760 samples received at GBRU for STEC testing, 1,002 non-O157 STEC cases were confirmed in England. Of the 1,002 non-O157 cases, 655 culture positive cases of 72 different serogroups were confirmed. For 21 isolates, a serotype could not be identified as the genes encoding the somatic O antigen did not match any known sequence in the database. Specimens for a further 347 cases in England were positive for stx genes on PCR at GBRU but an organism was not cultured (PCR positive-culture negative).
In Wales, 113 non-O157 cases of 40 different serotypes were confirmed and a further 66 were PCR positive-culture negative. The most common non-O157 serogroups isolated in 2019 were O26 (28/113, 25%), O146 (15/113, 13%), O128ab (10/113, 9%) and O91 (8/113, 7%) followed by O111 (4/113, 4%), O113 (4/113, 4%) and O156 (4/113, 4%).
Non-O157 STEC cases in England
In England, the 5 most common non-O157 serogroups isolated in 2019 were O26 (109/655, 17%) followed by O146 (98/655, 15%), O91 (81/655, 12%), O128ab (37/655, 6%) and O103 (28/655, 4%) (Table 3). The top 4 non-O157 serogroups were also most frequently detected in 2018. See full list of non-O157 STEC by stx type reported in 2019.
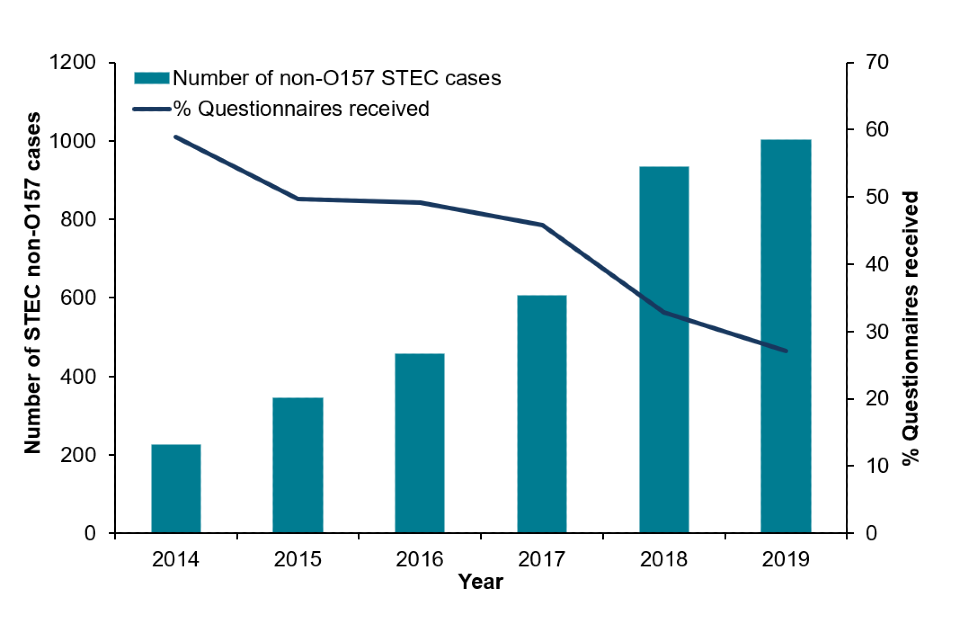
In 2019, ESQs were received for 27% (271/1,002) of STEC non-O157 cases, of which 80% (217/271) were for culture positive non-O157 STEC cases, and 16% of STEC cases confirmed only by PCR.
Since 2017, there has been decline in the number of ESQs received for STEC non-O157 cases, with a 41% reduction in the number of ESQs received from 2017 to 2019 (46% in 2017, 33% in 2018 and 27% in 2019), which may be associated with the implementation of algorithms to guide the local response of cases of STEC infection in 2018.
Of the 271 ESQs received for confirmed cases, 265 (98%) were symptomatic. Among the symptomatic cases, 93% (246/265) reported diarrhoea, including bloody diarrhoea in 51% (135/265) cases. This was accompanied by abdominal pain (183, 69%), nausea (105, 40%), fever (82, 31%), and vomiting (78, 29%). Eighty-five cases (32%) were hospitalised.
Figure 6: The number of confirmed STEC non-O157 cases and enhanced surveillance questionnaires received in England, 2019
HUS occurred in 22 confirmed and 1 probable STEC non-O157 cases. From the 23 cases of STEC non-O157 HUS, the following serogroups were isolated: O26 (n=8), O145 (n=4), O111 (n=1), O13-O135 (n=1), O146 (n=1), O45 (n=1), O80 (n=1) and O85 (n=1). Stx genes were detected in faecal samples for 4 cases, but no organism was isolated and antibodies to lipopolysaccharides of E.coli O26 was detected in a serum sample from one case.
Seven percent of confirmed STEC O26 cases developed HUS as compared to 3% in confirmed STEC O157 cases. HUS cases ranged from 5 months to 65 years of age and 56% (n=13) were between 1 and 4 years of age. There were 2 deaths reported among cases of non-O157 STEC.
ESQs were received for 63% (69/109) of all confirmed STEC O26 cases. Of the 67 cases that were symptomatic, all cases had diarrhoea, 69% (n=46) reported bloody diarrhoea, 72% (n=48) reported abdominal pain, 55% (n=37) reported nausea and 42% (n=28) reported vomiting. A higher proportion of STEC O26 cases (40%, n=27) reported hospitalisation compared to STEC O157 cases (29%, n=147).
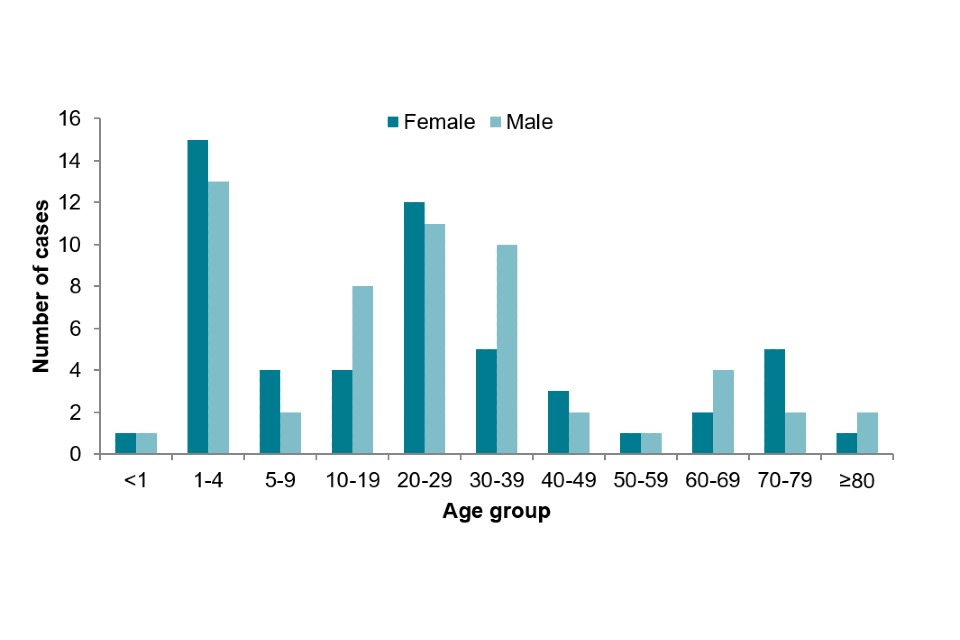
Of 109 STEC O26 cases in England, 53 (49%) were female. Children aged 1 to 4 years of age comprised 26% (n=28) of cases.
Figure 7: Age-sex distribution of STEC O26 cases in England, 2019
Among all cases of confirmed non-O157 STEC in 2019, 42% (n=424) possessed the Stx1 toxin alone, 29% (n=293) possessed Stx1 and Stx2 and 28% (n=285) possessed Stx2 toxin alone.
The most common stx subtypes detected through sequencing of the 655 isolates were stx1a only in 28% (n=185), stx1c only in 17% (n=114), stx1c stx2b in 13% (n=86), stx2b only in 10% (n=68), stx1a stx2b in 10% (n=66), stx2a only in 9% (n=60), stx1a stx2a in 4% (n=25), stx2d only in 2% (n=12), stx2c only in 1% (n=8), stx2f only in 1% (n=5), and stx2e only in 1% (n=4) (Table 3).
An additional 15 stx subtype combinations were detected in the samples of 21 cases, and for 1 case the stx subtype was not identified.
Table 3: Prevalence of stx subtype combination reported amongst the top 9 non-O157 STEC serogroups in England, 2019
| Serogroup | stx subtype | n | % |
|---|---|---|---|
| O26 | stx1a | 64 | 59 |
| stx2a | 28 | 26 | |
| stx1a stx2a | 15 | 14 | |
| stx2c | 2 | 1 | |
| O146 | stx1c | 38 | 39 |
| stx1c stx2b | 26 | 27 | |
| stx2b | 26 | 27 | |
| stx1a | 1 | 1 | |
| stx1a stx1c stx2b | 1 | 1 | |
| stx1a stx2b | 1 | 1 | |
| stx1c stx2c stx2b | 1 | 1 | |
| stx1c stx2d stx2b | 1 | 1 | |
| stx2b stx2d | 1 | 1 | |
| stx2c stx2d | 1 | 1 | |
| stx2d | 1 | 1 | |
| O91 | stx1a stx2b | 63 | 78 |
| stx2b | 11 | 14 | |
| stx1a | 6 | 7 | |
| stx2e | 1 | 1 | |
| O128ab | stx1a | 1 | 3 |
| stx1a stx2b | 1 | 3 | |
| stx1c | 6 | 16 | |
| stx1c stx2b | 17 | 46 | |
| stx2b | 12 | 32 | |
| O103 | stx1a | 25 | 89 |
| stx2a | 3 | 11 | |
| O113 | stx1c stx2b | 18 | 86 |
| stx2b | 1 | 5 | |
| stx2d | 2 | 10 | |
| O117 | stx1a | 21 | 95 |
| stx1a stx2a | 1 | 5 | |
| O76 | stx1c | 16 | 80 |
| stx1c stx2b | 3 | 15 | |
| stx2a | 1 | 5 | |
| O111 | stx1a | 15 | 88 |
| stx2a | 2 | 12 |
Outbreaks
STEC outbreaks investigated in England during 2019
Five STEC outbreaks affecting 65 people in England were investigated in 2019. Typically, an outbreak is investigated when an exceedance of 5 or more cases fall within the same 5 SNP single linkage cluster for clinically severe serotypes. However, an investigation may be undertaken after considering a number of factors, including the number of persons affected, temporal and geographic distribution, clinical severity, and the microbiological characteristics of the STEC strain.
Four outbreaks were caused by STEC serogroup O157, and thus 9% of confirmed STEC O157 cases were attributed to these 4 outbreaks (Table 4).
Despite epidemiological investigations, it was not possible to determine the vehicle and/or source of infection for outbreaks. There were 3 HUS cases associated with outbreaks in 2019, and no deaths.
Table 4: STEC O157 and non-O157 outbreaks in England, 2019
| Agent | Total number affected | Total laboratory confirmed | Hospitalised | Source | Region |
|---|---|---|---|---|---|
| STEC O157 PT 2 stx2a | 4 | 4 | 3 | Foodborne (suspected) | National |
| STEC O157 PT 21/28 stx2a stx2c | 10* | 10 | 4 | Foodborne | National |
| STEC O157 PT 2 stx2a | 7 | 7 | 4 | Outdoor event (suspected) | South East |
| STEC O157 PT 21/28 stx2a stx2c | 28† | 23 | 9 | Foodborne | Yorkshire and Humber |
| STEC O26 stx1a | 16‡ | 15 | 5 | Foodborne | National |
*14 cases total across the UK
†includes 7 cases affected and 2 confirmed in 2020
‡32 cases total across the UK
In October 2019, Public Health England (now UKHSA) and Public Health Scotland investigated an outbreak of STEC O26 stx1a eae positive. There were 31 laboratory-confirmed cases, of which 15 were resident in England (Table 2). This was the first non-O157 outbreak reported since the implementation of the Interim Public Health Operational Guidance for Shiga toxin producing Escherichia coli (STEC) algorithm.
The STEC operational guidance focuses on the presence of stx2a with the presence of eae and host factors for prioritisation of public health follow-up. Consequently, as the outbreak strain had stx1a only and cases were mostly healthy adults (median age of 28 years), many cases did not have ESQs completed when initially identified.
During this outbreak, there was a high number of cases reporting bloody diarrhoea (77%, n=10) and hospitalisation (38%, n=5) (based on English data only). This outbreak highlights the importance of stx1a in causing severe disease and is supported by increasing evidence in the literature that the presence of stx1a is also a risk factor for severe symptoms (5).
Conclusions
The number of STEC O157 cases in England continued to drop in 2019. The reason for this decline in recent years is unclear, although phage typing indicates a decrease in numbers of one of the most frequently detected types (PT 21/28) accompanied by a change in demography to older age groups (data not shown).
The detection of non-O157 STEC infections continued to rise in 2019 and reflects the more widespread use of PCR in frontline laboratories to detect a broader range of serogroups.
The most common non-O157 serogroup detected in 2019 was STEC O26 and cases infected with this serogroup had a higher proportion (7%) of HUS than O157 cases (3%).
The STEC O26 stx1a outbreak highlighted the importance of O26 serogroup in causing outbreaks and severe disease, as a public health concern.
UKHSA STEC publications in 2019
1. Adams N, Byrne L, Edge J, Hoban A, Jenkins C, Larkin L. Gastrointestinal infections caused by consumption of raw drinking milk in England & Wales, 1992–2017. Epidemiology and Infection. 2019;147
2. Adams NL, Byrne L, Rose TC, Adak GK, Jenkins C, Charlett A and others. Influence of socio-economic status on Shiga toxin-producing Escherichia coli (STEC) infection incidence, risk factors and clinical features. Epidemiology and Infection. 2019;147:e215
3. Awofisayo-Okuyelu A, Hall I, Arnold E, Byrne L, McCarthy N. Analysis of individual patient data to describe the incubation period distribution of Shiga-toxin producing Escherichia coli. Epidemiology and Infection. 2019;147:e162
4. Carroll KJ, Harvey-Vince L, Jenkins C, Mohan K, Balasegaram S. The epidemiology of Shiga toxin-producing Escherichia coli infections in the South East of England: November 2013-March 2017 and significance for clinical and public health. Journal of Medical Microbiology. 2019;68(6):930-9
5. Cowley LA, Dallman TJ, Jenkins C, Sheppard SK. Phage Predation Shapes the Population Structure of Shiga-Toxigenic Escherichia coli O157:H7 in the UK: An Evolutionary Perspective. Frontiers in Genetics. 2019;10(763):763
6. Elson R, Davies TM, Jenkins C, Vivancos R, O’Brien SJ, Lake IR. Application of kernel smoothing to estimate the spatio-temporal variation in risk of STEC O157 in England. Spatial and Spatio-temporal Epidemiology. 2020;32:100305
7. Greig DR, Jenkins C, Gharbia S, Dallman TJ. Comparison of single-nucleotide variants identified by Illumina and Oxford Nanopore technologies in the context of a potential outbreak of Shiga toxin-producing Escherichia coli. Gigascience. 2019;8(8)
8. Jenkins C, Dallman TJ, Grant KA. Impact of whole genome sequencing on the investigation of food-borne outbreaks of Shiga toxin-producing Escherichia coli serogroup O157:H7, England, 2013 to 2017. Eurosurveillance. 2019;24(4):1800346
9. Kintz E, Byrne L, Jenkins C, Mc CN, Vivancos R, Hunter P. Outbreaks of Shiga Toxin-Producing Escherichia coli Linked to Sprouted Seeds, Salad, and Leafy Greens: A Systematic Review. Journal of Food Protection. 2019;82(11):1950-8
10. Kirchner M, Sayers E, Cawthraw S, Duggett N, Gosling R, Jenkins C, and others. A sensitive method for the recovery of Escherichia coli serogroup O55 including Shiga toxin-producing variants for potential use in outbreaks. Journal of Applied Microbiology. 2019;127(3):889-96
11. Treacy J, Jenkins C, Paranthaman K, Jorgensen F, Mueller-Doblies D, Anjum M, and others. Outbreak of Shiga toxin-producing Escherichia coli O157:H7 linked to raw drinking milk resolved by rapid application of advanced pathogen characterisation methods, England, August to October 2017. Eurosurveillance. 2019;24(16):1800191
Acknowledgements
We are grateful to:
- the microbiologists, local authorities and local health protection and environmental health specialists who contribute data and reports to the NESSS
- the epidemiologists and information officers who have worked on NESSS
- staff in the GBRU for providing the Reference Laboratory Services and laboratory surveillance functions and expertise
- UKHSA Regional and Collaborating Public Health Laboratories and Food Water and Environmental Microbiology Services for providing a surveillance function for GI pathogens and testing of food and environmental samples during outbreak investigations
References
1. Launders, N. and others, Disease severity of Shiga toxin-producing E. coli O157 and factors influencing the development of typical haemolytic uraemic syndrome: a retrospective cohort study, 2009–2012. BMJ Open, 2016. 6(1): p. e009933
2. Dodd, C.C. and M.J. Cooper, Multidisciplinary response to the Escherichia coli 0104 outbreak in Europe. Mil Med, 2012. 177(11): p. 1406 to 10
3. Launders, N. and others, Outbreak of Shiga toxin-producing E. coli O157 associated with consumption of watercress, United Kingdom, August to September 2013. Eurosurveillance, 2013. 18(44)
4. Gobin, M. and others, National outbreak of Shiga toxin-producing Escherichia coli O157:H7 linked to mixed salad leaves, United Kingdom, 2016. Eurosurveillance, 2018. 23(18): p. 17-00197
5. Byrne, L., N. Adams and C. Jenkins, Association between Shiga Toxin-Producing Escherichia coli O157:H7 stx Gene Subtype and Disease Severity, England, 2009-2019. Emerging infectious diseases, 2020. 26(10): p. 2394 to 2400
6. Brandal, L.T. and others, Shiga toxin-producing escherichia coli infections in Norway, 1992-2012: characterization of isolates and identification of risk factors for haemolytic uremic syndrome. BMC Infectious Disease, 2015. 15: p. 324
7. Adams, N.L. and others, Shiga Toxin-Producing Escherichia coli O157, England and Wales, 1983-2012. Emerging infectious diseases, 2016. 22(4): p. 590 to 597
