Investigation into the risk to human health of avian influenza (influenza A H5N1) in England: technical briefing 5
Updated 14 July 2023
Applies to England
The UK Health Security Agency (UKHSA) is working with the Animal and Plant Health Agency (APHA), the Department for Environment, Food and Rural Affairs (Defra) and the public health agencies of the 4 nations to monitor the risk to human health of avian influenza (influenza A H5N1) in England. This briefing is produced to share data useful to other public health investigators and academic partners undertaking related work. It includes early evidence and preliminary analyses which may be subject to change.
Data reported in the technical briefing is as of 4 July 2023 (or as specified in the text) to allow time for analysis.
Summary
-
Detections of influenza in farmed poultry continue but remain at low levels compared to the last quarter of 2022. Wild bird detections continue to be geographically dispersed across England with a strong association with gull and tern species. The joint Defra and APHA assessment states that there continues to be a high level of influenza transmission in wild birds across the UK. No additional mammals have been received for testing since the last technical briefing.
-
In March 2023, asymptomatic surveillance of people exposed to avian influenza commenced. By 10 July 2023, 144 individuals from 8 infected premises have been tested, of which 4 were positive (2.7% positivity). This represents 2 additional detections since the last technical briefing. Since 2021, there have been 5 human influenza A(H5N1) detections in the UK in total, including a detection in December 2021 in a surveillance pilot.
-
The 2 new detections were in individuals exposed at 2 different premises to the previously reported detections. Detections are now associated with one backyard flock (2021), and 3 infected premises (2023 surveillance programme).
-
Detections in the asymptomatic surveillance study are assessed for immediate public health action based on the timing of positivity related to exposure. The first new detection is difficult to interpret due to lack of information on sample timing and may be consistent with infection or contamination of the respiratory tract. The second new detection is likely to represent contamination. Precautionary contact tracing was undertaken.
-
One partial viral sequence from first of the new detections, and one full viral sequence from the second of the new detections is available, but no viral sequences from birds on the linked infected premises are available to date. Both these human detection sequences are influenza A(H5N1) clade 2.3.4.4b and consistent with the UK genotype AIV48, also known as the A/gull/France/22P015977/2022-like genotype.
-
Globally, since December 2021, 15 human detections of influenza A(H5N1) have been reported officially. Twelve of these cases are from clade 2.3.4.4b. This includes the 2 new detections in England since the last technical briefing. There is no evidence of human-to-human transmission from these cases.
-
International surveillance of mammals varies but continues to show spill-over into mammalian species. Since the last technical briefing, there are reports of detections of influenza A(H5N1) in cats in Poland and of a continued sea lion die-off in South America. Genomes available show changes associated with mammalian adaptation in PB2. There is no evidence confirming sustained mammalian transmission. Detailed investigations and increased numbers of genomic sequences from such outbreaks and nearby avian infections would be informative.
-
There is no evidence of human-to-human transmission from any detections and recent findings do not change the assessment of human health risk, which remains at level 3 (limited mammalian transmission, low confidence) as described in the previous technical briefing (see also the qualitative assessment on influenza A(H5N1) infections in non-avian UK wildlife from the multi-agency Human Animal Infections and Risk Surveillance (HAIRS) Group).
Part 1. Asymptomatic influenza surveillance of people exposed to infected birds
1.1 Surveillance update
As of 10 July 2023, 202 individuals were identified as having been within the biosecurity area on the 8 included sites. Of these, 144 (71%) participated in the surveillance programme. Participants include farm staff, staff involved in the culling operation, veterinarians and other health and safety staff.
The study protocol detailing the study design and procedures was recently published.
For the purposes of immediate public health response, test results are divided into those that are of uncertain significance and those that may be representative of true infection. Positive tests taken either during the period of exposure or within 48 hours of exposure may reflect nasopharyngeal contamination rather than true infection and are therefore classified as of uncertain significance.
A positive detection taken at least 48 hours after the last exposure is classified as possible infection and managed as such on a precautionary basis. This assessment is a low confidence assessment but can be used to support immediate public health decision making. Further assessment is undertaken through confirmatory testing, repeat sampling, and serology.
1.2 Surveillance results
A total of 144 individuals have consented to participate; 83 (57.6%) staff directly involved in the culling operation, 30 (20.8%) farm staff, 11 (7.6%) veterinarians, and 20 (13.9%) others such as health and safety managers and volunteers.
Four human detections of influenza A(H5N1) clade 2.3.4.4b have been confirmed from 3 geographically distinct sites in 2023. Genomic analysis of all 4 detections are consistent with or confirmed as genotype AIV48.
For human detections in the UK, UKHSA will use the nomenclature UK/Year/ Number. This will include all detections regardless of whether they are judged to be contamination or infection.
Detection UK/2021/001 was from December 2021 and was part of a pilot asymptomatic surveillance program. The further detections (UK/2023/001-004) were all detections from the current study. Detections UK/2023/001 and UK/2023/002 were reported in technical briefing 4.
Paired sera were obtained from 2 of the 5 total UK detections since December 2021, and either acute or convalescent serum was obtained from the other 3 detections but may be challenging to interpret. Analysis is underway.
Summarised data on detections of influenza A(H5N1) viral nucleic acid in samples taken from humans in 2023 is shown in Table 1. Samples were reported as influenza A positive in 2 UKHSA laboratories and were confirmed using 2 independent H5-specific assays (Spackman and colleagues, 2002; Slomka and colleagues, 2007).
Table 1. Summary data on human influenza A(H5N1) UK detections 2023.
Day 0 is the day of recruitment (during farm work) on which an observed swab is taken. Exposure may finish on day 0 or may continue usually for a short period. The participant is requested to self swab on days 2, 5 and 8.
| Detection | First day of exposure* | Last day of exposure* | Day of positive samples* | Day of first negative sample after testing positive* | Symptoms | Rapid assessment (PCR only) | UK Genotype | Virus culture |
|---|---|---|---|---|---|---|---|---|
| UK/2023/001 | -5 | 2 | 0 | 2 | No | Uncertain significance, likely contamination | AIV48† | Negative |
| UK/2023/002 | 0 | 2 | 4 and 5 | 13 | No | Possible infection | AIV48† | Negative |
| UK/2023/003 | -1 | 2 | Unclear**(either 2, 5 or 8) | Unclear** | Sore throat, myalgia*** | Uncertain significance | AIV48† | Negative |
| UK/2023/004 | 0 | 0 | 0 | 2 | No | Uncertain significance, likely contamination | AIV48 | In progress |
*The day of recruitment is considered day 0.
**UK/2023/003 returned self taken swabs without dates.
*** Symptoms are not definitively associated with time of positivity.
† Partial match due to incomplete sequence data.
1.3 Contact tracing
UK/2023/003
All close household contacts (3) remained clinically asymptomatic. 29 other contacts from the infected premises were followed up as part of the study and tested negative.
UK/2023/004
All close household contacts (3) remained clinically asymptomatic. 20 other contacts from the infected premises were followed up as part of the study and tested negative.
1.4 Genomics
Viral genome sequencing was undertaken on human detection UK/2023/003 and sequence data for 6 of 8 segments was generated (Global Initiative on Sharing All Influenza Data (GISAID) accession EPI_ISL_17980947). Sequence was not generated for PB1 and PB2. Sequencing of avian samples from the farm is currently underway.
Viral genome sequencing was also undertaken on human detection UK/2023/004, with sequence data generated for all 8 segments (sequence data will be uploaded to GISAID in due course).
The HA segment for both human detections were identified as clade 2.3.4.4b. The genotype for detection UK/2023/003 could not be confirmed as all 8 segments were not sequenced successfully. The available segments were most closely related to UK genotype AIV48, also known as the A/gull/France/22P015977/2022-like genotype. The genotype for detection UK/2023/004 was confirmed as AIV48.
Genotype AIV48 was first detected in the UK in June 2022, with further detection across Great Britain and Crown Dependencies since then. In England, this includes 26 poultry cases, 3 captive or rehabilitated birds, 45 wild birds and 2 mammals (foxes) for which sequence data is available.
The genome sequence for both detections were compared to the first AIV48 genome detected in the UK (EPI_ISL_13782459: A/H5N1 A/chicken/England/085598/2022) to identify amino acid substitutions within the available segments. Non-synonymous mutations for both sequences are listed in Table 2. There are 12 non-synonymous changes in the sequence data available for human detection UK/2023/003 compared to the AIV48 reference sequence. All these mutations have been observed in at least 1 other UK AIV48 genome.
There are 26 non-synonymous changes in the sequence data available for human detection UK/2023/004. Of these, 10 are in segments that were not successfully sequenced in detection UK/2023/003 and 10 are shared with detection UK/2023/003. There are 6 non-synonymous changes in detection UK/2023/004 that have not been observed in any UK AIV48 genomes to date. Variation is expected within each genotype.
Table 2. Mutational profile of sequencing detections UK/2023/003 and UK/2023/004 relative to AIV48 reference (EPI_ISL_13782459: A/chicken/England/085598/2022)
Dashes (-) indicate insufficient coverage at that position.
| Protein or segment | Amino acid Position | AIV48 reference | Detection UK/2023/003 | Detection UK/2023/004 | Percentage (%) of UK AIV48 sequences with mutation |
|---|---|---|---|---|---|
| PB2 | 187 | K | - | Q | 0 |
| PB2 | 443 | K | - | N | 23 |
| PB2 | 619 | L | - | M | 23 |
| PB2 | 631 | M | - | V | 21 |
| PB2 | 685 | R | - | G | 24 |
| PB2 | 699 | K | - | R | 57 |
| PB1 | 172 | D | - | E | 2.3 |
| PB1 | 175 | N | - | D | 80 |
| PB1 | 181 | I | - | M | 79 |
| PB1 | 648 | A | - | S | 56 |
| PA | 30 | I | I | N | 0 |
| PA | 94 | I | V | V | 23 |
| PA | 204 | R | R | K | 0 |
| PA | 211 | M | I | I | 23 |
| PA | 315 | F | L | L | 23 |
| PA | 350 | S | G | S | 1.2 |
| PA | 404 | A | V | V | 23 |
| HA | 87* | T | I | I | 98 |
| HA | 104 | D | G | G | 59 |
| HA | 143 | T | T | A | 0 |
| HA | 298 | V | I | I | 23 |
| NP | 452 | R | R | K | 0 |
| NA | 79 | A | D | D | 12 |
| NA | 136 | Q | H | H | 22 |
| NA | 394 | V | I | V | 1.2 |
| M2 (MP) | 68 | P | P | S | 23 |
| NS1 (NS) | 55 | E | E | G | 0 |
| NS1 (NS) | 180* | I | V | V | 98 |
*This is a mutation that has been identified in the AIV48 reference only and not in other AIV48 genomes.
Part 2. Avian infections
2.1 Current epidemiological situation
The dominant pathotype circulating in avian species across England continues to be high pathogenicity avian influenza A(H5N1).
Avian epidemiology
From 1 October 2022, APHA has confirmed HPAI influenza A(H5N1) in poultry species at 157 premises in England, and in wild birds from 592 sites in England. Further information on the latest avian influenza situational update in England is published on GOV.UK.
The number of detections at infected premises remains relatively low compared to the period before the housing order was implemented. This housing order has now been lifted. The frequency of detections of avian influenza in wild birds is relatively low compared to the levels observed in the last quarter of 2022 but have maintained their geographical spread across England (Figures 1a and 1b). The joint Defra and APHA assessment is that there continues to be a high level of influenza transmission in wild birds across the UK.
Figure 1a. Confirmed detections of avian influenza at infected premises by quarter in England from 1 October 2022 to 4 July 2023. Data provided by APHA
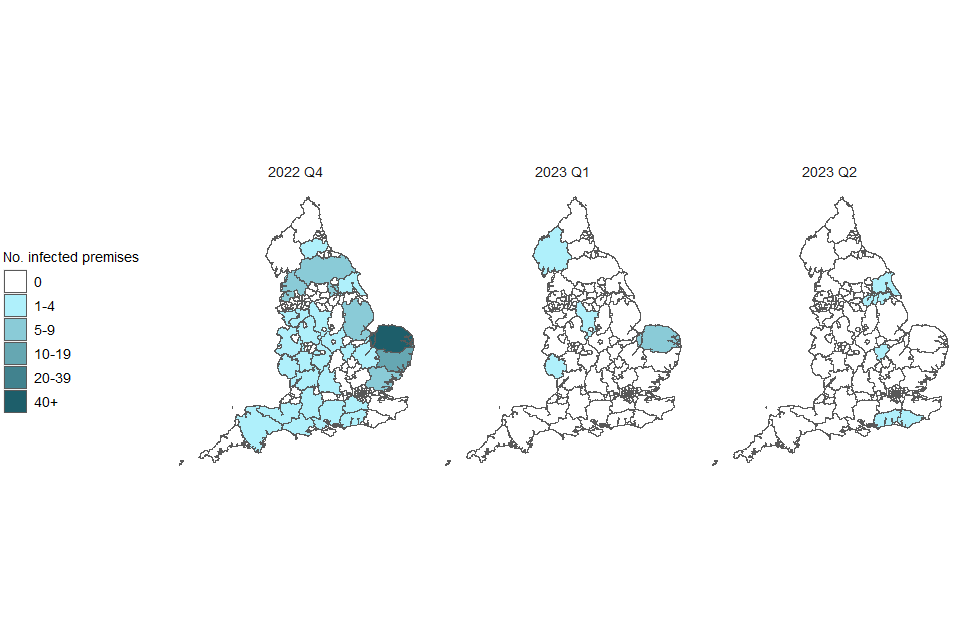
Figure 1b. Confirmed detections of avian influenza in wild birds by quarter in England from 1 October 2022 to 4 July 2023. Data provided by APHA
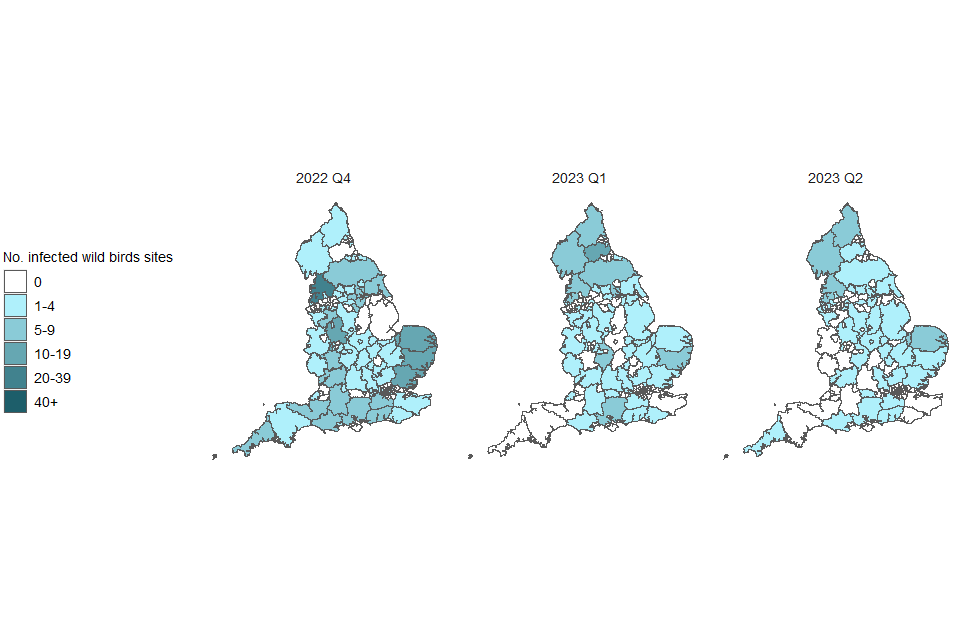
These maps contain National Statistics data © Crown copyright and database right 2022.
Q4 (2022) = Quarter 4 (1 October 2022 to 31 December 2022)
Q1 (2023) = Quarter 1 (1 January 2023 to 31 March 2023)
Q2 (2023) = Quarter 2 (1 April 2023 to 4 July 2023)
Supplementary data is not available for these figures to prevent deductive disclosure.
Part 3. Mammalian infections
Since the previous technical briefing no wild mammals reported to the competent authority met the testing criteria for submission to the national reference laboratory for testing. Previously, testing of mammalian samples was triaged on a case-by-case basis. However, in April 2023 a guidance document that defines the suspect case definition and diagnostic testing criteria for influenza A(H5N1) infection in mammals was published by Defra and APHA.
APHA surveillance has detected influenza A(H5) in 23 of 247 wild mammals collected since October 2021. Of these, 8 samples tested positive through reactive testing whilst 18 positives were detected in material submitted retrospectively for testing.
There are continued reports of detections in mammals internationally (Figure 2). The sampling frame is unknown and there is no standardised approach to surveillance or reporting.
Figure 2. International reports of mammalian influenza A(H5N1) detections collated through epidemic intelligence surveillance from 1 January 2021 to 4 July 2023

The data used in Figure 2 can be found in the accompanying spreadsheet. Data is sourced from the Emerging Infections and Zoonoses epidemic intelligence scanning database from official and unofficial reports (including media reports), which may include a small number of duplicate entries due to incomplete information. Event date indicates collection date where known, or notification date when collection date unknown.
Since the last technical briefing, there have been reports of detections of influenza A(H5N1) in cats in Poland, and a continued sea lion die-off in South America. Known mammalian adaptation mutations are seen in the PB2 segments of available genomes in both situations.
As of 12 July 2023, sequence data from 104 mammalian cases of A(H5N1) (including human) have been uploaded to GISAID with a collection date after 1 July 2022. Of those, 97 have a complete PB2 sequence. Further analysis of these genomes is being undertaken by UKHSA to assess mutations previously reported to have an association with mammalian adaptation.
Potential human exposures
Human exposures to avian influenza undergo a risk assessment by UKHSA Health Protection Teams (HPTs). These teams manage and record exposures in the outbreak information system HPZone, as detailed in previous technical briefings.
Since 1 October 2022, 2,577 exposure episodes across England were entered into HPZone, including a further 93 since the last technical briefing. Guidance on interpreting HPZone data has been published in previous technical briefings.
Information is also collected by HPTs using surveillance forms, as described in technical briefing 1, with caveats on completeness and lag. Between 1 October 2022 and 4 July 2023, information was returned for 315 (42.0%) out of 749 incidents this season and includes records of 3376 exposures (individuals may be recorded in more than one event if they are exposed multiple times). Over half (63%) of the surveillance forms received from HPTs relate to wild bird incidents (201 out of 315 forms returned). Wild bird incidents often involve fewer exposed individuals than farmed poultry outbreaks of avian influenza.
Personal protective equipment (PPE) use was reported in 782 (23.16%) exposures. Antiviral prophylaxis was reported for 313 (9.27%) exposures. For individuals reporting flu-like symptoms, 52 symptomatic swabs were taken and tested (73.2% of those eligible in this category). All tests were reported as negative for influenza A(H5).
3.2 Human cases
Globally, 15 human cases of influenza A(H5N1) have been reported officially since December 2021. No human-to-human transmission has been reported related to these cases.
Cases from influenza A(H5N1) clade 2.3.4.4b
Since the last technical briefing, 2 cases of influenza A(H5N1) clade 2.3.4.4b have been reported from England as detailed above in section 1. Details on other cases can be found in previous technical briefings. A total of 12 human cases of influenza A(H5N1) clade 2.3.4.4b have been reported since December 2021.
UKHSA continues to carry out horizon scanning for epidemiological reports relevant to emerging influenza in humans and animals.
3.3 Influenza samples referred to UKHSA for characterisation
UKHSA receives influenza positive clinical samples referred from NHS and regional public health laboratories (PHLs) for whole genome sequencing, virus isolation and antigenic characterisation, year-round. This is described in further detail in technical briefing 1. The sensitivity of this system for detecting emerging viruses is currently under assessment.
No unsubtypeable influenza samples were received since 1 June 2023.
Sources and acknowledgments
Data sources
Data relating to animal health surveillance and investigations taking place across England was obtained from the APHA. This includes data from wild bird surveillance, notifiable disease reports at infected premises and detections in mammals.
Data on international reports of mammals is sourced from the UKHSA Emerging Infections and Zoonoses epidemic intelligence scanning database from official and unofficial reports (including media reports).
Surveillance forms are completed by UKHSA HPTs for each confirmed setting (includes both poultry and wild bird settings). This includes the follow-up of exposed persons and details of exposure. Data is enhanced with laboratory records for respiratory testing held by UKHSA.
Details of exposed individuals are also collected from HPZone, the UKHSA case management system.
International surveillance data of human cases of avian influenza is reported by the World Health Organization (WHO) under the International Health Regulations and routinely collated by UKHSA.
Authors of this report
Rachel Abbey, Wendy Barclay, Neil Bray, Ian Brown, Ashley Banyard, Alexander Byrne, Fernando Capelastegui, Lorenzo Cattarino, Meera Chand, Neil Cunningham, Richard Puleston, Eileen Gallagher, Natalie Groves, Berkin Hack, Katja Hoschler, Susan Hopkins, Kate Howell, Joe James, Angie Lackenby, Anissa Lakhani, Anika Singanayagam, Nick Watkins, Maria Zambon.
Contributors
- UKHSA Acute Respiratory Infections
- UKHSA Data, Analytics and Surveillance Team
- UKHSA Data Science and Geospatial team
- UKHSA Genomics Public Health Analysis
- UKHSA Rapid Investigation Team
- UKHSA Respiratory Virus Unit
- UKHSA Emerging Infections and Zoonoses Team
- Animal and Plant Health Agency
- Imperial College London
- Francis Crick Institute
- The Pirbright Institute
Avian Influenza Technical Group
The Avian Influenza Technical Group includes members with expertise in clinical infectious diseases, clinical research, epidemiology, genomics and virology:
- Meera Chand (Chair), UKHSA
- Ashley Banyard, APHA
- Wendy Barclay, Imperial College London
- Ian Brown, APHA
- Alexander Byrne, APHA
- Andre Charlett, UKHSA
- Fergus Cumming, UKHSA
- Neil Ferguson, Imperial College London
- Eileen Gallagher, UKHSA
- Natalie Groves, UKHSA
- Yper Hall, UKHSA
- Bassam Hallis, UKHSA
- Susan Hopkins, UKHSA
- Katja Hoschler, UKHSA
- Munir Iqbal, The Pirbright Institute
- Joe James, APHA
- Rowland Kao, University of Edinburgh
- Angie Lackenby, UKHSA
- Nicola Lewis, Francis Crick Institute
- Nicholas Loman, UKHSA and University of Birmingham
- Paul Millar, Health and Social Care Northern Ireland
- Thomas Peacock, Imperial College London
- Richard Puleston, UKHSA
- Oliver Pybus, Royal Veterinary College and University of Oxford
- Andrew Rambaut, University of Edinburgh
- Helen Roberts, Defra
- Anika Singanayagam, UKHSA
- Nick Watkins, UKHSA
- Christopher Williams, Public Health Wales
- Anthony Wilson, Food Standards Agency
- Maria Zambon, UKHSA
Acknowledgements
The authors are grateful to those teams and groups providing data for these analyses including:
- Animal and Plant Health Agency
- Pirbright Institute
- Imperial College London
