Investigation into the risk to human health of avian influenza (influenza A H5N1) in England: technical briefing 1
Updated 14 July 2023
Applies to England
The UK Health Security Agency (UKHSA) is working with the Animal and Plant Health Agency (APHA) and the Department for Environment, Food and Rural Affairs (Defra) to investigate the risk to human health of avian influenza (influenza A H5N1) in England. This briefing is produced to share data useful to other public health investigators and academic partners undertaking related work. It includes early evidence and preliminary analyses which may be subject to change.
Data reported in the technical briefing is as of 13 December 2022 (or as specified in the text) to allow time for analysis.
Levels of human health risk related to the outbreak of avian influenza in England
These risk levels were developed by the Technical Group to help to establish triggers for enhancing assessment and response. The avian influenza outbreak can be considered to fall into one of 6 potential levels of transmission.
Level 0 (Baseline)
Avian influenza circulating in birds within normal bounds of prevalence and with normal epidemiological dynamics.
Level 1
Avian influenza circulating in birds with altered epidemiological dynamics and/or increased prevalence.
Level 2
Level 1 plus detection of spillover into mammals.
Level 3
Evidence of viral genomic changes that provide an advantage for mammalian infection.
Level 4
Sustained transmission in non-human mammalian species or any human detection and mutations in haemagglutinin (HA) which allow transmission. (A single human detection in a person exposed to infected birds, without HA mutations, does not raise the risk level to 4.)
Level 5
Any human-to-human transmission.
The UK risk is currently assessed as at level 3.
Main data points
Since 1 October 2022, the start of the current reporting year for avian influenza, APHA has notified UKHSA that highly pathogenic avian influenza (HPAI) A(H5N1) has been confirmed in avian species at 130 premises in England. Wild bird testing is undertaken on a geographically representative sample of birds, with 447 influenza A (H5N1) detections at 280 locations in England reported since 1 October 2022. Since the introduction of the poultry housing order on 7 November 2022, numbers of infected premises are decreasing but detections in wild birds continue to suggest high levels of circulating virus in the UK.
From 1 October 2022 to 15 December 2022, health protection systems have recorded 2,085 human exposure episodes (where a person was directly exposed to an infected bird). There is likely to be substantial under ascertainment.
Detailed data on incidents (health protection responses to avian influenza detections) is incomplete. Based on the 29% of incidents for which there is data (1 October 2022 to 13 December 2022), personal protective equipment (PPE) was used in 27.3% of exposures, and antiviral prophylaxis in 15.9% of exposures. Symptoms were reported following 31 (4.3%) exposures, with 24 symptomatic swabs being carried out (77.4% of those eligible). There have been no detections of avian influenza viruses in humans the UK during the current reporting year (from 1 October 2022 to date) and there was one human detection in the UK in the preceding reporting year (1 October 2021 to 30 September 2022).
APHA report that 20 mammals have been retrospectively tested, of which 8 were influenza A (H5N1) positive. Four of these have genome sequences available and all show the presence of a mutation which is associated with potential advantages for mammalian infection. This is very limited data but, together with international data, is suggestive of sporadic mammalian spillover events.
Some clinical and regional public health laboratories undertake influenza subtyping and refer influenza A which is unsubtypable by standard clinical assays to UKHSA for characterisation. From 1 January 2022 to 8 December 2022, 44 samples were referred to UKHSA as unsubtypable and of these 18 were seasonal H1 or H3 viruses, 11 had no virus detected, 6 had a low viral load precluding further characterisation, and 9 are still being characterised. Assessment of the sensitivity and completeness of this system for the detection of novel influenza viruses is being undertaken.
Part 1. Risk assessment as of 13 December 2022
This assessment is based on reports made from APHA and other partners to UKHSA. Data sharing is being established but UKHSA has not viewed the current data in full.
UK virus population
There is an increase in confirmed cases of influenza A infected birds (high confidence). In 2022, there has been year-round maintenance of influenza infection in indigenous wild birds which represents a change compared to the usual seasonal pattern in which infections die out over the summer. Compared to the previous risk assessment of 11 November 2022, there are a reducing number of infected premises following the introduction of the national housing order for farmed poultry, but still high levels of detections in dead wild birds.
Influenza A H5N1 is the predominant influenza virus subtype detected in wild birds and farmed flocks in the UK (high confidence). There is diversity within the UK population of H5N1 viruses with 11 genotypes detected since October 2021, including some reassortment with low pathogenic avian influenza viruses (LPAIVs). However, 7 of these genotypes have constituted a limited number of detections. The dominant circulating genotypes since October 2021 are AIV09 and AIV07-B2. Since October 2022, AIV09 is the predominant genotype. Another currently detected genotype in poultry is AIV48 which includes genes from gull-associated influenza viruses.
Genomic surveillance is proportionate for poultry outbreaks (a genome is generated for every affected premise). There is a limited genomic surveillance sampling in wild birds. APHA select birds to test and report that testing is distributed in time and space with host species consideration. There is very limited surveillance of mammals. Genomic data lags 7 to 10 days behind date of sample collection for poultry and currently longer for mammals.
Extent of human exposure in the UK
Owing to the disease burden in birds there is an increased interface between humans and infected birds (high confidence). In particular the high number of wild birds and domestic flocks with influenza A infection, especially where personal protective equipment is not worn, increases the likelihood of human exposures to this virus (moderate confidence).
Propensity to cause mammalian and human infection
Available surveillance data reported by APHA do not suggest widespread mammalian adaptation of this virus (low to moderate confidence).
Mutations known to be advantageous in mammalian infections are infrequent in the available genomic data from avian viruses however these data are lagging. APHA report that there is evidence of direct spill over from birds into some ‘scavenger’ wild mammalian species within the UK (and others noted outside the UK). In the UK 8 mammals, out of a total of 20 targeted from samples collected during 2021 to 2022 and retrospectively tested by APHA, were positive for influenza A (H5N1).
The species affected (foxes and otters) are presumed to have direct high-level exposure to infected birds based on feeding behaviour and food preferences. The 4 available influenza genomes from these positive mammals all show the PB2 E627K substitution. This mutation is known to be acquired rapidly after infection of a mammalian host in some influenza viruses and is associated with enhanced polymerase activity.
The rapid and consistent acquisition of the PB2 mutation in mammals may imply this virus has a propensity to cause zoonotic infections and further assessment should be made of the properties of this mutation. There is also recent confirmed transmission of a virus similar to the AIV48 genotype between mink in Spain, but the published genomes available show no evidence of significant HA mutation.
There is incomplete genotype to phenotype understanding and genomic data must be supplemented by in vitro and animal model studies.
There have been 4 instances of influenza A H5N1 2.3.4.4b detection in humans (1 UK, 1 USA, 2 Spain) between December 2021 and December 2022. There is limited asymptomatic testing of human contacts of bird cases in the UK and international surveillance is variable. Nevertheless, by comparison with other zoonotic infections including influenza viruses, these data suggest that zoonotic infections are infrequent (low confidence).
Ability to cause (a) severe infection and (b) asymptomatic infection in humans
There are no detected severe human cases associated with Influenza A H5N1 (clade 2.3.4.4b) in the UK or internationally. There is insufficient information to judge the risk of asymptomatic or mild disease due to limited testing in human contacts of infected birds.
Human-to-human transmission
There is no evidence of sustained human to human transmission (moderate to high confidence). Subtyping surveillance in the NHS or through NHS referral to UKHSA is incomplete and could delay detection. There is insufficient information to assess the occurrence of limited human to human transmission such as transmission within households.
The current H5N1 2.3.4.4b viruses in UK birds react well against antisera raised against an available Influenza A(H5) World Health Organization (WHO) candidate vaccine virus (CVV) (A/Astrakhan/3212/2020), developed for pandemic preparedness and coordinated by WHO.
Assessment
The avian influenza outbreak in the UK is assessed as at risk level 3 although there is limited mammalian surveillance data. At present, there are no indicators of increasing risk to human health, however this is a low confidence assessment. The risk assessment is dynamic and requires regular review during this period of unusually high levels of transmission in birds with mammalian spillover. In vitro and animal model data are required. Enhancements to mammalian and human asymptomatic infection surveillance are both in preparation.
Part 2. Epidemiology update
2.1 Current epidemiological situation
There have been unprecedented levels of avian influenza circulating in England over 2021 and 2022. The dominant subtype currently circulating in avian species across England is highly pathogenic avian influenza (HPAI) A(H5N1).
Since 1 October 2022, the start of the current 2022 to 2023 seasonal period, detections of avian influenza have been confirmed in both wild birds and infected premises which can include domestic flocks and poultry farms. APHA undertake diagnostic testing in any flocks where notifiable avian disease cannot be excluded by official veterinarians. The APHA also undertake passive surveillance of wild bird populations from the notification of mortality events by members of the public reported through a Defra helpline. Testing of avian influenza in wild birds is subject to dynamic surveillance policies and thresholds for collection based on resource implications and the current situation regarding infection trends across the UK.
Since the start of the 2022 to 2023 season, the APHA has notified UKHSA that HPAI A(H5N1) has been confirmed in avian species at 130 premises in England. Animal health surveillance has also detected A(H5N1) in 447 wild birds from 280 locations in England. One detection of A(H6N2) avian influenza was confirmed at an infected premises in the North West of England.
Detections of avian influenza at infected premises have continued through the reporting period and have fluctuated during this time (Figure 1). A national housing order for poultry was introduced on 7 November 2022. Wild bird detections have continued to demonstrate that the background risk from avian influenza in wild birds remains very high (Figure 1).
Detections of avian influenza have been concentrated in the East of England and the East Midlands, driven in particular by high numbers of infected premises in these areas (Figures 1 and 2). However, detections in wild birds have been more widespread across the country (Figure 1).
Figure 1. Confirmed detections of avian influenza in poultry and wild birds by setting in England from 1 October 2022 to 13 December 2022. Data provided by APHA
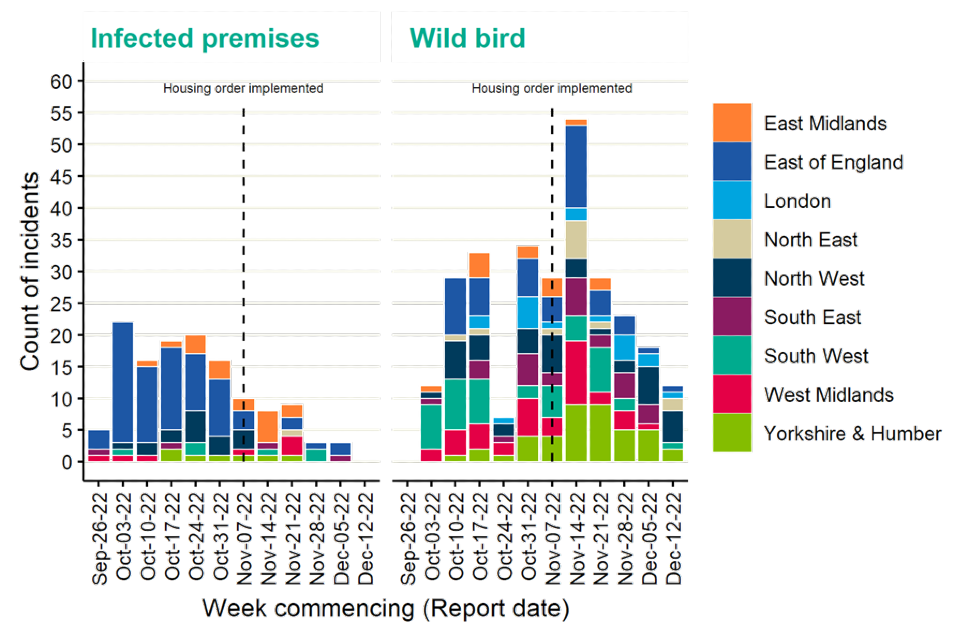
The data used in this graph can be found in the accompanying spreadsheet.
Figure 2. Choropleth map of avian influenza incidents detected in poultry and wild birds across England by upper tier local authority (UTLA), from 1 October 2022 to 13 December 2022
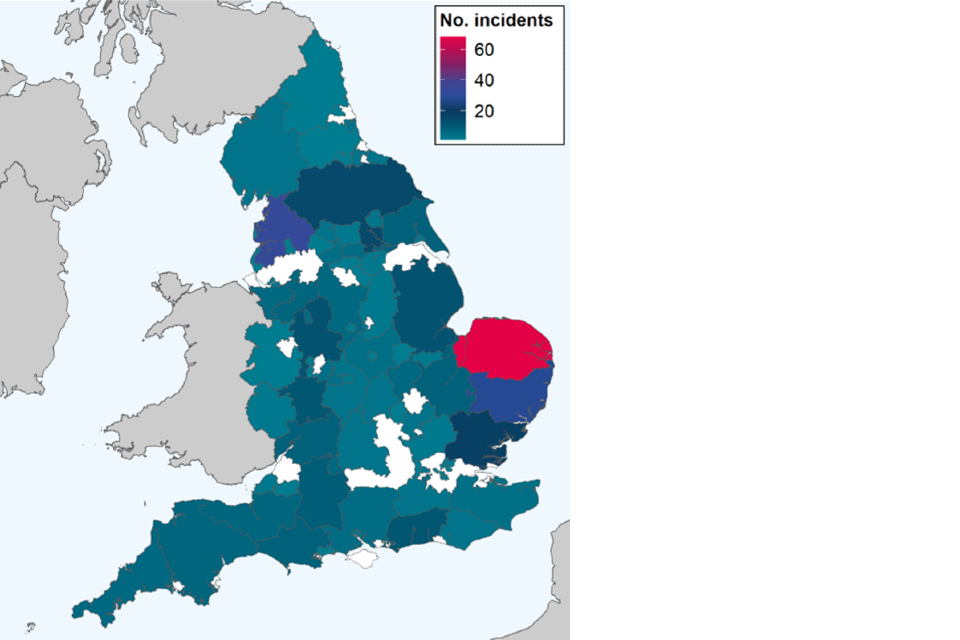
This map contains National Statistics data © Crown copyright and database right 2022.
Data source: Animal and Plant Health Agency 2022.
Supplementary data is not available for this figure.
The APHA has undertaken retrospective testing of stored mammalian samples that were found either linked to infected premises or as having unusual clinical behaviours (for example, neurological signs), raising suspicion of infection with avian influenza. This has led to the identification of A(H5N1) infection in 8 out of 20 mammals collected since December 2021. Of these, 4 were from England and were detected in foxes, and 4 detections in Scotland, all of which were in otters.
2.2 Summary of exposed persons
The increase in detections of avian influenza has led to human exposures relating to farmed and wild birds, which are managed by UKHSA health protection teams (HPTs). Workers on farms can usually be easily identified, but the extent of human exposures to infected wild birds is more difficult to determine and there is likely to be substantial under ascertainment.
Public health guidance advises the use of personal protective equipment and antiviral prophylaxis for individuals at risk of exposure to avian influenza. Post-exposure, individuals are offered antiviral prophylaxis with a follow-up or health monitoring period of up to 10 days.
Symptomatic individuals are referred for swabbing to detect possible infection. There is also an option for asymptomatic swabbing for those eligible (individuals who did not wear PPE at the time of exposure but who remain symptom free during the follow-up period) as part of a surveillance pilot. However, the uptake from individuals for this pilot continues to be extremely low.
Details of exposed individuals are recorded on the HPZone case management system at UKHSA. HPZone was interrogated from 1 October 2022 to 15 December 2022. Data was selected for analysis where the infectious agent was ‘avian influenza’.
Over the reporting period, 2,085 exposure episodes were entered into the HPZone system. Exposed individuals were mainly identified as male (71.5%). Females accounted for 19.5% and gender was unknown for 9% of exposure episodes. Distribution of gender was similar across all age ranges (Figure 3). Age was unknown for 33.1% of exposure episodes entered into HPZone.
Figure 3. Age-sex distribution of exposure episodes captured in HPZone from 1 October 2022 to 15 December 2022
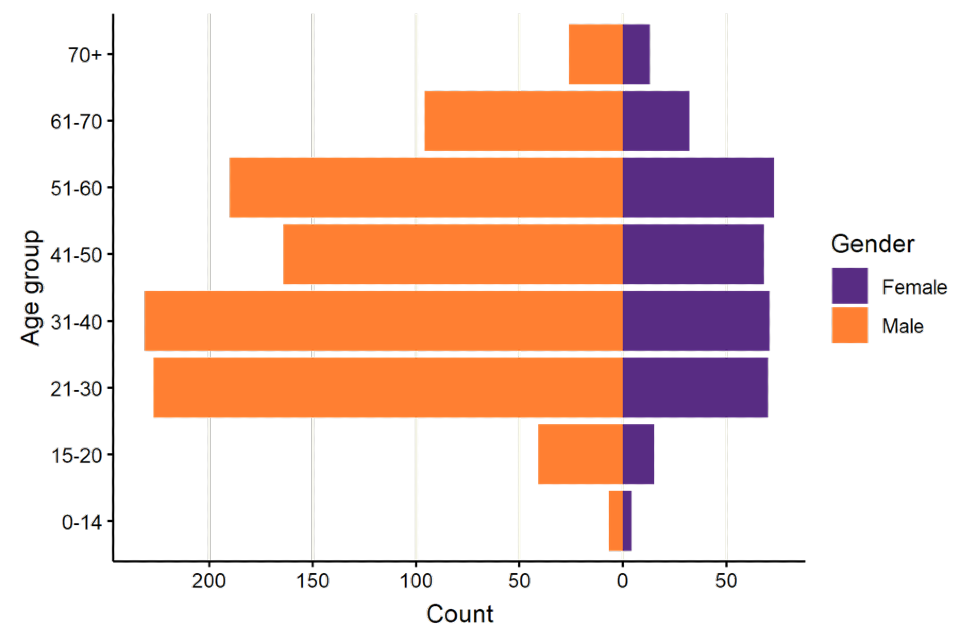
The data used in this graph can be found in the accompanying spreadsheet.
Following the epidemiology of avian influenza detections, the distribution of exposed individuals is concentrated in the East of England and the East Midlands (Figure 4).
Figure 4. Exposures reported on HPZone from 1 October 2022 to 15 December 2022
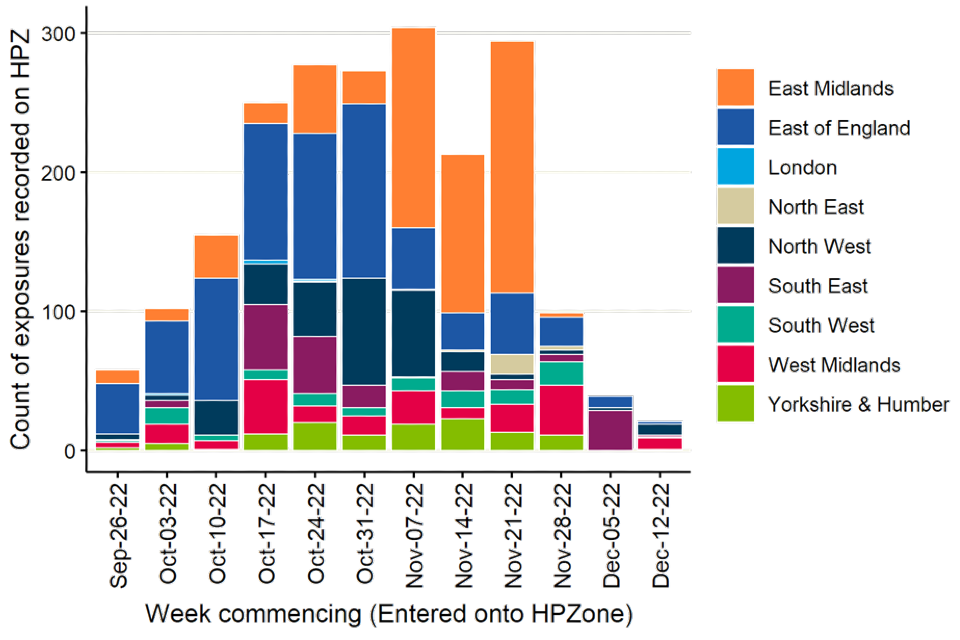
The data used in this graph can be found in the accompanying spreadsheet.
Information is also collected by HPTs using surveillance forms for each situation to document PPE use, antiviral use, symptom status, and any swabbing. These forms are returned to the national epidemiology team and linked to laboratory records held by UKHSA on respiratory testing.
Between 1 October 2022 and 13 December 2022, information was returned for 119 (29%) out of 411 incidents this season and recorded 716 exposure events. Individuals may be recorded in more than one event if they are exposed multiple times (exposure episodes are differentiated by time and location).
Data for other incidents is unavailable at present, however, in regions with high avian influenza activity, HPTs prioritise public health action over data collection and reporting.
The majority of surveillance forms received from HPTs relate to wild bird incidents (90 out of 119 forms returned) comprising 75.6% of data. Wild bird incidents often involve fewer exposed individuals.
Analysis of the available data indicates that PPE use was reported in 268 (37.4%) exposures. Antiviral prophylaxis was reported for 114 (15.9%) exposures. Symptoms were reported following 31 (4.3%) exposures, with 24 symptomatic swabs being carried out (77.4% of those eligible). Thirteen asymptomatic swabs were performed and reported as part of an enhanced surveillance pilot from consenting individuals.
Data should be interpreted with caution due to the incomplete nature of information collected from exposed individuals. Data receipt is expected to lag to allow for adequate follow-up time of exposed persons to elapse.
There were no human detections of influenza H5 in England during the 2022 to 2023 season.
In December 2021, one human case of A(H5N1) was confirmed in England in an exposed person who remained asymptomatic throughout. However, this detection was in the context of close and prolonged exposure to infected poultry and contaminated material, without PPE. (Oliver and colleagues 2022)
In addition to the human case reported from England, 3 human cases of A(H5N1) of the same clade (A(H5N1) 2.3.4.4.b) were reported internationally by the WHO between December 2021 and December 2022. This includes one case from the USA and 2 cases from Spain. All 3 cases were involved in poultry decontamination and culling activities. The case from USA reported mild fatigue, and the 2 cases from Spain were asymptomatic. No human-to-human transmission has been reported.
UKHSA continues to carry out horizon scanning for epidemiological reports relevant to emerging influenza in humans and animals.
2.3 Capability to detect human cases of H5N1
Weekly national flu and COVID-19 reports are published by UKHSA. These include a detailed breakdown of influenza virus characterisation.
UKHSA receives influenza positive clinical samples referred from NHS and regional public health laboratories (PHLs) for whole genome sequencing, virus isolation and antigenic characterisation, year-round. This provides a picture of circulating influenza viruses in the community and in hospital settings. This system is designed to understand which viruses are causing seasonal influenza and is not calibrated to detect small numbers of novel influenza cases.
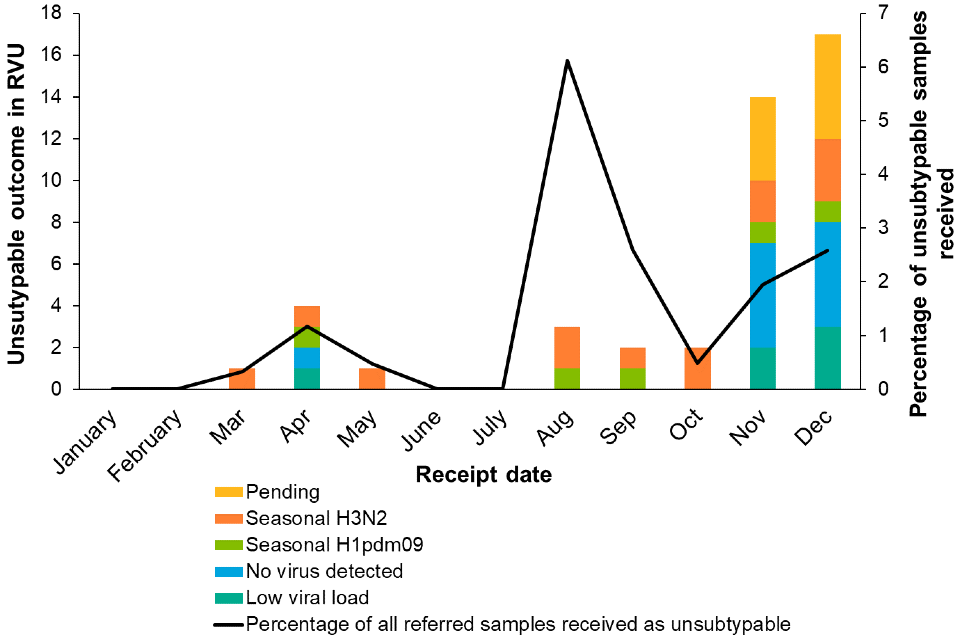
UKHSA request that samples detected as influenza A positive in NHS or UKHSA PHLs which are not assigned a subtype through routine assays are forwarded to the UKHSA Respiratory Virus Unit (RVU). The UKHSA RVU performs subtyping by polymerase chain reaction (PCR) and genome sequencing. The sensitivity of this system for detecting emerging viruses is currently under assessment.
Figure 5 shows the results for unsubtypable samples referred in 2022 up to 8 December 2022. As of 8 December 2022, 1.3% of samples (n=44) referred to UKHSA Colindale from NHS or UKHSA PHLs in 2022 were influenza A unsubtypable. Of these, 41% (n=18) were characterised as seasonal H1 or H3 viruses, with 25% (n=11) having no virus detected and 14% (n=6) as having detectable but insufficient viral load to achieve a subtyping result. Characterisation is ongoing for 9 samples.
Figure 5. Influenza A unsubtypable samples referred in 2022 up to 8 December 2022
The data used in this graph can be found in the accompanying spreadsheet.
Part 3. Genomic surveillance
The current genomic analysis is performed by APHA. UKHSA has requested full data access to undertake human health risk assessment in parallel.
APHA has published a pre-print describing the diversity of H5 avian influenza genomes in the UK between 2020 and 2022. The paper describes the genomic diversity sequences between October 2020 and May 2022, as well as the prevalence of some characterised mammalian adaptation mutations. Between October 2020 and October 2021, there were a range of H5Nx infections detected, with all highly pathogenic avian influenza virus (HPAIV) haemagglutinin (HA) sequences belonging to clade 2.3.4.4b. However, when typed by the HA segment, the majority of sequences between October 2021 and May 2022 were H5N1 (195 out of 196 sequences). One avian sequence in this dataset contained the mutation E627K in the polymerase (PB2) protein, which may confer advantages for mammalian infection.
From October 2021 to May 2022, no further avian sequences were found to contain the E627K substitution. In total across the whole period of genomic surveillance of the current influenza A (H5N1) outbreak, APHA report that they have assed 457 influenza genomes from birds for this mutation and detected it only once.
However, APHA have reported that all 4 H5N1 sequences obtained from mammals (foxes and otters) did possess this substitution.
Part 4. Planned rapid laboratory assessments and early data
4.1 Candidate vaccine viruses assessment
The development of influenza candidate vaccine viruses (CVVs), coordinated by WHO, remains an essential component of the overall global strategy for influenza pandemic preparedness.
This assessment was undertaken by the Worldwide Influenza Centre at the Francis Crick Institute as part of the existing UK commitment to supply data to the WHO global influenza programme and is based on haemagglutination inhibition assay data generated by UK and international laboratories in support of the programme.
The virus detected in the UK lies within the H5 clade 2.3.4.4b which is now widespread across Africa, Asia, Europe and North America. However, there is diversity within this clade and viruses from some countries, including in Eastern Europe, West Africa, Cambodia and Vietnam, are less well recognised by antiserum raised against the A/Astrakhan/3212/2020 2.3.4.4b CVV. As a result of this antigenic drift in some geographic areas, a recommendation was made in September to add a second 2.3.4.4b CVV recommendation (A/chicken/Ghana/AVL-76321VIR7050-39/2021-like). Recent antigenic analyses of H5 clade 2.3.4.4b viruses isolated from birds in the UK have all been genetically and antigenically similar to the original A/Astrakhan/3212/2020 CVV.
Although the recommended CVV is a good antigenic match for currently circulating viruses in the UK, partners within this expert technical group will continue to characterise emerging strains, both genetically and antigenically, within poultry and those that might be detected within humans.
4.2 Antivirals assessment
Three influenza specific treatments are approved by the Medicines and Healthcare products Regulatory Agency (MHRA) in the UK. Two neuraminidase inhibitors (NAIs), oseltamivir and zanamivir, are already deployed, and one cap-dependent endonuclease inhibitor (baloxavir marboxil) is not yet marketed in the UK. Evidence suggests that all 3 drugs have activity against influenza A(H5N1).
UKHSA undertakes routine genomic surveillance for antiviral resistance in seasonal influenza viruses, using established databases of mutations associated with reduced drug susceptibility or reduced inhibition by NAIs including among avian influenza viruses, and will extend this to avian viruses routinely once data are made available.
Analysis performed by APHA on 316 full genome A(H5N1) HPAI virus sequences obtained from poultry and wild birds in the UK and Crown Dependencies from October 2021 to October 2022 did not identify any amino acid substitutions in NA associated with NAI reduced inhibition. A single viral sequence with an I38T amino acid substitution was identified. This substitution is known to reduce susceptibility to baloxavir in human seasonal A(H3N2) and A(H1N1)pdm09 viruses.
Phenotypic NAI susceptibility testing requires virus isolation and is performed as an enzyme inhibition assay at the Francis Crick Institute. Phenotypic testing for baloxavir susceptibility is being developed at the Crick Institute.
APHA, UKHSA and the WHO Collaborating Centre at the Francis Crick Institute will collaborate going forwards over avian influenza antiviral susceptibility surveillance.
Part 5. Further planned work
5.1 Preliminary knowledge gaps assessment
UKHSA is currently undertaking a research and evidence gaps analysis relating to the avian influenza outbreak in the UK with internal and external stakeholders. A group of experts were convened to discuss the emerging priority gaps for research, evaluation, and surveillance studies. Further workshops are planned to refine and prioritise these gaps and develop research questions. UKHSA will work with stakeholders, including academic partners and national research funders, to identify active research studies in these areas and develop and implement studies to address remaining gaps.
5.2 Surveillance
Mammalian surveillance
APHA are developing a surveillance pipeline for submission and testing of mammalian samples. Carcasses may be submitted where animals are:
- found dead in areas located near to defined infected premises, or
- are displaying clinical signs that may be considered indicative of potential infection with HPAIV
These surveillance activities are primarily driven through the Diseases of Wildlife Scheme.
Enhancement of asymptomatic surveillance in humans
UKHSA is developing protocols for intensive sampling of individuals working on sentinel infected premises. These can be used to establish human asymptomatic infection parameters at baseline and in response to changes in viral or epidemiological features of the outbreak.
5.3 Improvements to data and analysis
The following areas have been identified for improvements to surveillance, data and analytics:
1. UKHSA should receive full genomic sequencing data in real time from APHA and undertake continuous human health focused genomic risk assessment.
2. Current influenza surveillance systems, which are primarily tailored towards monitoring seasonal influenza, should be assessed for sensitivity and timeliness in detecting emerging influenza viruses.
3. A more detailed assessment is needed of the wild bird sampling framework and consideration should be given to:
- reporting more detailed metrics including positivity rate, species and numbers of reported but untested birds
- assessing whether more detailed sampling studies could improve understanding of the viral population and transmission in the UK
Sources and acknowledgments
Data sources
Data relating to animal health surveillance and investigations taking place across England obtained from the APHA. This includes data from wild bird surveillance, notifiable disease reports at infected premises and detections in mammals.
Surveillance forms are completed by UKHSA HPTs for each confirmed setting (includes both poultry and wild bird settings). This includes the follow-up of exposed persons and details of exposure. Data is enhanced with laboratory records for respiratory testing held by UKHSA.
Details of exposed individuals are also collected from HPZone, the UKHSA case management system.
International surveillance data of human cases of avian influenza is reported by the WHO under the International Health Regulations and routinely collated by UKHSA.
Authors of this report
Rachel Abbey, Carolina Arevalo, Ashley Banyard, Wendy Barclay, Ian Brown, Alexander Byrne, Fernando Capelastegui, Lorenzo Cattarino, Meera Chand, David Edwards, Eileen Gallagher, Irene Gonsalvez, Katja Hoschler, Susan Hopkins, Munir Iqbal, Joe James, Angie Lackenby, Nicola Lewis, Thomas Peacock, Richard Puleston, Jess Tarrant, Nick Watkins, Maria Zambon, Anissa Lakhani.
Contributors
- UKHSA Data Science and Geospatial team
- UKHSA Genomics Public Health Analysis
- UKHSA Respiratory Virus Unit
- UKHSA Research and Evaluation
- UKHSA Research Support and Governance Office
- UKHSA Rapid Investigation Team
- Animal and Plant Health Agency
- Imperial College London
- Francis Crick Institute
- The Pirbright Institute
Avian Influenza Technical Group
The Avian Influenza Technical Group includes members with expertise in clinical infectious diseases, clinical research, epidemiology, genomics and virology:
- Meera Chand (Chair), UKHSA
- Wendy Barclay, Imperial College London
- Alexander Byrne, APHA
- Ashley Banyard, APHA
- Ian Brown, APHA
- Neil Ferguson, Imperial College London
- Yper Hall, UKHSA
- Bassam Hallis, UKHSA
- Susan Hopkins, UKHSA
- Katja Hoschler, UKHSA
- Munir Iqbal, The Pirbright Institute
- Joe James, APHA
- Angie Lackenby, UKHSA
- Nicola Lewis, Francis Crick Institute
- Nicholas Loman, UKHSA and University of Birmingham
- Berit Mueller-Pebody, UKHSA
- Derren Ready, UKHSA
- Thomas Peacock, Imperial College London
- Richard Puleston, UKHSA
- Andrew Rambaut, University of Edinburgh
- Nick Watkins, UKHSA
- Maria Zambon, UKHSA
- Esther Robinson, UKHSA
Acknowledgements
The authors are grateful to those teams and groups providing data for these analyses including:
- Animal and Plant Health Agency
- Francis Crick Institute
- Pirbright Institute
- Imperial College London
